Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for ETV3
Z-value: 4.55
Transcription factors associated with ETV3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV3
|
ENSG00000117036.12 | ETV3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV3 | hg38_v1_chr1_-_157138474_157138555 | 0.44 | 1.7e-11 | Click! |
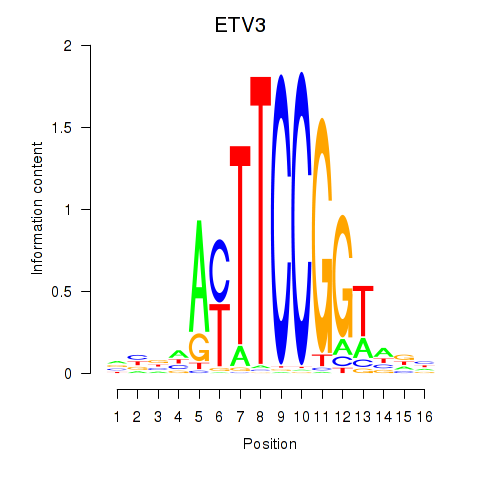
Activity profile of ETV3 motif
Sorted Z-values of ETV3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 27.3 | GO:0001555 | oocyte growth(GO:0001555) regulation of progesterone secretion(GO:2000870) |
| 5.0 | 15.0 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 4.9 | 19.7 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 4.4 | 13.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 3.0 | 8.9 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 2.8 | 11.3 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 2.6 | 7.9 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 2.6 | 7.8 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 2.4 | 7.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 2.2 | 4.3 | GO:0090182 | regulation of secretion of lysosomal enzymes(GO:0090182) |
| 2.1 | 10.5 | GO:0035900 | response to isolation stress(GO:0035900) |
| 2.1 | 6.2 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 1.9 | 36.7 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 1.9 | 5.7 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 1.8 | 23.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 1.8 | 5.3 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 1.7 | 8.6 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 1.7 | 5.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 1.7 | 15.1 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 1.6 | 8.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 1.6 | 1.6 | GO:1990910 | response to hypobaric hypoxia(GO:1990910) |
| 1.6 | 12.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 1.5 | 7.6 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 1.4 | 4.2 | GO:0070079 | peptidyl-lysine hydroxylation to 5-hydroxy-L-lysine(GO:0018395) histone arginine demethylation(GO:0070077) histone H3-R2 demethylation(GO:0070078) histone H4-R3 demethylation(GO:0070079) |
| 1.3 | 6.5 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 1.3 | 9.0 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 1.3 | 2.5 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 1.3 | 11.3 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 1.2 | 6.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 1.2 | 15.8 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 1.2 | 6.1 | GO:0060296 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 1.2 | 2.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 1.1 | 4.5 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 1.1 | 3.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 1.1 | 4.4 | GO:0016559 | peroxisome fission(GO:0016559) |
| 1.0 | 7.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 1.0 | 3.1 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 1.0 | 9.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 1.0 | 8.6 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 1.0 | 13.4 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.9 | 3.8 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.9 | 6.6 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.9 | 8.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.9 | 5.4 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.9 | 7.3 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.9 | 6.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.8 | 10.8 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.8 | 2.5 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.8 | 4.9 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.8 | 4.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.8 | 7.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.8 | 7.2 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.8 | 3.2 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.8 | 8.6 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.7 | 2.2 | GO:0035048 | splicing factor protein import into nucleus(GO:0035048) |
| 0.7 | 3.6 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.7 | 2.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.7 | 2.9 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.7 | 10.7 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.7 | 4.3 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.7 | 17.0 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.7 | 7.6 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.7 | 2.0 | GO:0090521 | regulation of embryonic cell shape(GO:0016476) glomerular visceral epithelial cell migration(GO:0090521) |
| 0.7 | 3.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.7 | 1.3 | GO:0018963 | phthalate metabolic process(GO:0018963) |
| 0.6 | 6.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.6 | 28.6 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.6 | 7.7 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.6 | 8.7 | GO:0075713 | establishment of integrated proviral latency(GO:0075713) |
| 0.6 | 1.7 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.6 | 7.8 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.5 | 27.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.5 | 4.7 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.5 | 7.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.5 | 5.4 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.5 | 6.7 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.5 | 7.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.5 | 32.8 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.5 | 8.9 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.5 | 11.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.5 | 12.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.4 | 4.0 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.4 | 3.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.4 | 6.5 | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) |
| 0.4 | 4.4 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.4 | 6.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.4 | 2.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.4 | 3.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.4 | 2.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.4 | 4.1 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.4 | 15.3 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.4 | 11.5 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.4 | 5.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.3 | 1.3 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.3 | 5.9 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.3 | 9.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.3 | 2.8 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.3 | 9.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.3 | 3.2 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.3 | 4.4 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.3 | 12.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.3 | 3.6 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.3 | 6.0 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.3 | 16.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.3 | 7.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.3 | 3.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.3 | 2.6 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.3 | 2.3 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.3 | 5.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.3 | 5.8 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.2 | 5.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 7.9 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.2 | 2.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 1.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.2 | 6.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.2 | 8.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 0.2 | GO:1903004 | regulation of protein K63-linked deubiquitination(GO:1903004) positive regulation of protein K63-linked deubiquitination(GO:1903006) |
| 0.2 | 5.7 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.2 | 4.0 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.2 | 2.8 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.2 | 8.9 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.2 | 7.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 2.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 2.9 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.2 | 11.2 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.2 | 3.0 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.2 | 1.3 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 7.3 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.2 | 1.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 3.5 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.2 | 2.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 23.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.2 | 3.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.2 | 1.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 2.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 7.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.2 | 4.4 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 | 1.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 3.9 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 7.1 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 6.3 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 2.8 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 2.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 3.8 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 0.1 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) positive regulation of mast cell cytokine production(GO:0032765) |
| 0.1 | 2.6 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 2.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.6 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 1.0 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.5 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 1.7 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.1 | 0.4 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.1 | 0.5 | GO:0046543 | thelarche(GO:0042695) development of secondary female sexual characteristics(GO:0046543) mammary gland branching involved in thelarche(GO:0060744) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.7 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 2.8 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 2.0 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 0.5 | GO:0051306 | metaphase/anaphase transition of mitotic cell cycle(GO:0007091) regulation of mitotic sister chromatid separation(GO:0010965) mitotic sister chromatid separation(GO:0051306) |
| 0.1 | 1.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.6 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 4.5 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 5.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.7 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 1.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 2.6 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.1 | 1.7 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 2.0 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 3.1 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 1.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 2.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.7 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.6 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.5 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.6 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 1.3 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 2.1 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.7 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 2.1 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 4.1 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 1.5 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.3 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 1.6 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 | 1.3 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 4.9 | GO:0072659 | protein localization to plasma membrane(GO:0072659) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 1.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 1.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.0 | GO:0055028 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 3.3 | 16.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 2.9 | 8.6 | GO:0044609 | DBIRD complex(GO:0044609) |
| 2.8 | 11.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 2.7 | 10.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 2.5 | 12.3 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 2.3 | 15.8 | GO:1990745 | EARP complex(GO:1990745) |
| 2.1 | 17.1 | GO:0033647 | host cell cytoplasm(GO:0030430) host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) host cell cytoplasm part(GO:0033655) |
| 1.8 | 7.1 | GO:0000801 | central element(GO:0000801) |
| 1.7 | 8.7 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 1.7 | 8.6 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 1.6 | 4.9 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 1.4 | 8.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 1.4 | 12.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 1.3 | 3.9 | GO:0005745 | m-AAA complex(GO:0005745) |
| 1.3 | 6.5 | GO:0032044 | DSIF complex(GO:0032044) |
| 1.2 | 7.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 1.1 | 5.7 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 1.1 | 6.8 | GO:0070545 | PeBoW complex(GO:0070545) |
| 1.1 | 5.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 1.0 | 11.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 1.0 | 5.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 1.0 | 5.7 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.9 | 5.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.9 | 4.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.9 | 12.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.8 | 2.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.8 | 2.5 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.8 | 8.8 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.8 | 7.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.8 | 6.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.7 | 18.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.7 | 6.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.7 | 19.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.6 | 7.8 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.6 | 19.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.6 | 7.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.5 | 4.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.5 | 3.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.5 | 3.8 | GO:0070187 | telosome(GO:0070187) |
| 0.5 | 7.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.4 | 7.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.4 | 8.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.4 | 11.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.4 | 3.7 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.4 | 5.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.4 | 4.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.4 | 4.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.4 | 1.5 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.3 | 3.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.3 | 3.9 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 3.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 1.6 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.2 | 28.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.2 | 3.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 19.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.2 | 2.0 | GO:0044447 | axoneme part(GO:0044447) |
| 0.2 | 28.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 2.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 5.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 13.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 1.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 24.0 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 2.0 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.2 | 1.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 5.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 1.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 5.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 2.8 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 1.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 6.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.7 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 9.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 8.9 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 4.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 2.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 3.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 4.0 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 3.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 3.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 7.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 2.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 6.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 20.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 4.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 9.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 4.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 5.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 3.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 2.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 22.4 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 19.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 19.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 4.1 | 12.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 4.1 | 36.7 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 2.8 | 11.3 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 2.6 | 10.5 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 2.2 | 8.9 | GO:0032810 | sterol response element binding(GO:0032810) |
| 1.8 | 5.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 1.6 | 4.9 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 1.6 | 9.7 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 1.5 | 7.6 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.5 | 4.4 | GO:0052816 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 1.4 | 4.2 | GO:0033746 | histone demethylase activity (H3-R2 specific)(GO:0033746) histone demethylase activity (H4-R3 specific)(GO:0033749) |
| 1.3 | 11.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 1.3 | 6.3 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 1.2 | 12.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 1.2 | 15.7 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 1.2 | 6.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 1.2 | 15.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 1.1 | 20.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 1.1 | 5.7 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 1.1 | 3.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 1.1 | 12.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 1.1 | 4.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 1.1 | 28.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 1.0 | 8.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.9 | 7.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.9 | 6.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.9 | 5.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.9 | 4.3 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.9 | 2.6 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.9 | 8.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.8 | 7.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.7 | 6.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.7 | 9.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.7 | 3.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.6 | 15.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.6 | 17.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.6 | 4.4 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.6 | 15.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.6 | 1.7 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.6 | 5.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.5 | 5.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.5 | 1.6 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.5 | 2.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.5 | 12.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.4 | 1.3 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.4 | 1.3 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.4 | 4.3 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.4 | 2.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.4 | 3.4 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 27.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.4 | 3.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.4 | 5.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.4 | 2.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.4 | 7.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 1.9 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.4 | 3.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.4 | 6.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.4 | 2.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.3 | 6.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 8.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.3 | 7.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 6.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 5.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.3 | 7.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 7.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 58.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.3 | 12.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.3 | 6.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 2.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 4.8 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 3.0 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.3 | 5.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 2.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.3 | 4.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 1.1 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.2 | 6.1 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.2 | 1.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 3.6 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.2 | 2.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 4.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 16.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 1.4 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.2 | 1.7 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 1.0 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.2 | 12.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.2 | 0.5 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 4.9 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 10.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 8.5 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.1 | 3.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 6.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 9.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.9 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 14.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 6.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 6.4 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 1.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 4.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 4.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 2.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 4.5 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 22.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 3.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 5.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 1.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.6 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 27.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 12.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 2.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 8.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 7.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 3.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 6.1 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 1.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 30.0 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 3.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 17.9 | GO:0016491 | oxidoreductase activity(GO:0016491) |
| 0.0 | 2.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 5.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 4.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 2.1 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 13.4 | GO:0016773 | phosphotransferase activity, alcohol group as acceptor(GO:0016773) |
| 0.0 | 1.5 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.1 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 11.9 | GO:0003677 | DNA binding(GO:0003677) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 15.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 8.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.3 | 17.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.3 | 5.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.3 | 23.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 13.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 6.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 18.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 7.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 17.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 12.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 10.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 7.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 3.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 15.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 9.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 2.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 6.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 9.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 5.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.1 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 5.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 5.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 32.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 3.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 4.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 3.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.6 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 15.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.7 | 10.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.7 | 7.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.6 | 9.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.6 | 7.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.6 | 11.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.5 | 11.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.5 | 3.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.5 | 8.7 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.4 | 8.8 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.4 | 5.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.4 | 29.1 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.3 | 14.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 4.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 4.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 11.8 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.3 | 7.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 20.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 4.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 17.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 5.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 6.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 5.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 2.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 26.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.2 | 2.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 3.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 5.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 13.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 2.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 4.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 4.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 4.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 6.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 2.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 7.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 2.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 7.2 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 3.3 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 2.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |