Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
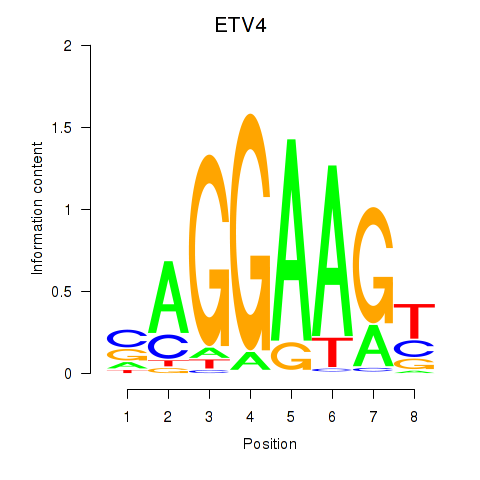
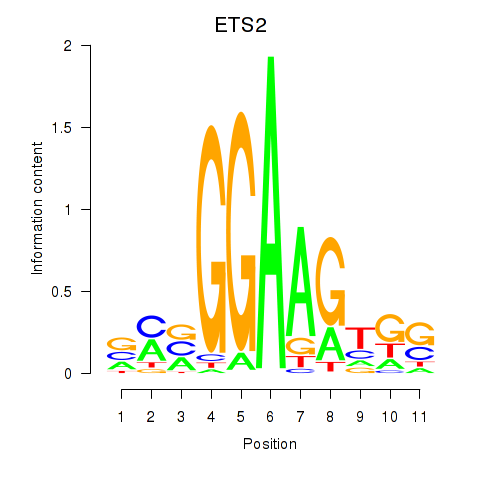
Results for ETV4_ETS2
Z-value: 2.82
Transcription factors associated with ETV4_ETS2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV4
|
ENSG00000175832.13 | ETV4 |
|
ETS2
|
ENSG00000157557.13 | ETS2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV4 | hg38_v1_chr17_-_43545707_43545732 | 0.23 | 6.8e-04 | Click! |
| ETS2 | hg38_v1_chr21_+_38805968_38805983, hg38_v1_chr21_+_38805895_38805951, hg38_v1_chr21_+_38805165_38805217 | 0.07 | 3.2e-01 | Click! |
Activity profile of ETV4_ETS2 motif
Sorted Z-values of ETV4_ETS2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV4_ETS2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.3 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 4.6 | 13.7 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 4.3 | 12.9 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 4.1 | 20.3 | GO:2000870 | oocyte growth(GO:0001555) regulation of progesterone secretion(GO:2000870) |
| 4.0 | 16.0 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 3.7 | 18.4 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 3.1 | 9.2 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 3.0 | 9.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 3.0 | 8.9 | GO:0072683 | T cell extravasation(GO:0072683) |
| 2.9 | 8.7 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 2.9 | 8.6 | GO:0070079 | peptidyl-lysine hydroxylation to 5-hydroxy-L-lysine(GO:0018395) histone arginine demethylation(GO:0070077) histone H3-R2 demethylation(GO:0070078) histone H4-R3 demethylation(GO:0070079) |
| 2.8 | 5.6 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 2.6 | 18.1 | GO:0033590 | response to cobalamin(GO:0033590) |
| 2.6 | 28.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 2.5 | 10.2 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 2.5 | 10.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 2.5 | 9.8 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 2.4 | 7.2 | GO:0050748 | negative regulation of lipoprotein metabolic process(GO:0050748) |
| 2.3 | 11.3 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 2.1 | 2.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 2.1 | 26.7 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 1.7 | 5.2 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 1.7 | 10.2 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 1.7 | 14.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 1.6 | 6.6 | GO:1904378 | maintenance of unfolded protein(GO:0036506) protein localization to cytosolic proteasome complex(GO:1904327) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 1.6 | 4.8 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 1.6 | 6.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 1.6 | 7.9 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 1.6 | 4.7 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 1.6 | 4.7 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 1.5 | 4.6 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 1.5 | 7.5 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 1.5 | 4.5 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 1.5 | 4.5 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 1.4 | 4.2 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 1.4 | 4.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 1.3 | 1.3 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 1.3 | 4.0 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 1.3 | 4.0 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 1.3 | 1.3 | GO:1990910 | response to hypobaric hypoxia(GO:1990910) |
| 1.3 | 11.7 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 1.3 | 12.8 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 1.3 | 8.8 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 1.3 | 17.5 | GO:0016559 | peroxisome fission(GO:0016559) |
| 1.2 | 5.0 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 1.2 | 2.5 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 1.2 | 6.1 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 1.2 | 9.7 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 1.2 | 10.9 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 1.2 | 3.6 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 1.2 | 8.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 1.2 | 3.5 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 1.2 | 3.5 | GO:0034092 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of telomeric DNA binding(GO:1904742) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 1.2 | 6.9 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 1.1 | 8.0 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 1.1 | 7.9 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 1.1 | 2.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 1.1 | 1.1 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 1.1 | 4.3 | GO:0071879 | UDP-glucose catabolic process(GO:0006258) positive regulation of adrenergic receptor signaling pathway(GO:0071879) negative regulation of type B pancreatic cell development(GO:2000077) |
| 1.1 | 6.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 1.1 | 3.2 | GO:0035048 | splicing factor protein import into nucleus(GO:0035048) |
| 1.1 | 3.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 1.0 | 3.1 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) |
| 1.0 | 4.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 1.0 | 14.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 1.0 | 5.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 1.0 | 20.2 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 1.0 | 4.0 | GO:0021539 | subthalamus development(GO:0021539) |
| 1.0 | 3.9 | GO:0090301 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 1.0 | 5.8 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.0 | 2.9 | GO:0019089 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 0.9 | 3.8 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.9 | 27.3 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.9 | 4.7 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.9 | 2.8 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) relaxation of skeletal muscle(GO:0090076) |
| 0.9 | 2.8 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.9 | 2.8 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.9 | 4.6 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.9 | 4.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.9 | 4.6 | GO:1901910 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.9 | 9.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.9 | 15.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.9 | 0.9 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.9 | 5.3 | GO:0032252 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.9 | 2.7 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.9 | 2.6 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.9 | 4.4 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.9 | 11.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.9 | 8.6 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.9 | 1.7 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) |
| 0.9 | 6.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.8 | 2.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.8 | 5.9 | GO:0030421 | defecation(GO:0030421) |
| 0.8 | 3.2 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.8 | 8.9 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.8 | 4.9 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.8 | 3.2 | GO:1902473 | regulation of protein localization to synapse(GO:1902473) |
| 0.8 | 2.4 | GO:0072716 | response to actinomycin D(GO:0072716) |
| 0.8 | 0.8 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.8 | 7.2 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.8 | 3.2 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.8 | 3.1 | GO:0044407 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.8 | 9.3 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.8 | 4.6 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.8 | 10.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.7 | 9.6 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.7 | 1.5 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.7 | 2.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.7 | 1.4 | GO:1900238 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) negative regulation of phospholipid biosynthetic process(GO:0071072) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.7 | 18.9 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.7 | 5.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.7 | 2.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.7 | 2.6 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.7 | 2.6 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.6 | 7.7 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.6 | 2.6 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.6 | 17.7 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.6 | 1.9 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) regulation of embryonic cell shape(GO:0016476) |
| 0.6 | 6.9 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.6 | 6.9 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.6 | 1.9 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.6 | 2.5 | GO:0086053 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.6 | 4.9 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.6 | 10.4 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.6 | 7.3 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.6 | 6.7 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.6 | 3.6 | GO:0001692 | histamine metabolic process(GO:0001692) histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.6 | 3.5 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.6 | 3.5 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.6 | 3.5 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.6 | 1.8 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.6 | 18.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.6 | 1.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.6 | 4.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.6 | 6.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.6 | 1.7 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.6 | 1.7 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.6 | 3.4 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.6 | 3.9 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.6 | 5.6 | GO:0060717 | chorion development(GO:0060717) |
| 0.6 | 1.7 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.6 | 1.7 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.6 | 1.1 | GO:0061141 | lung ciliated cell differentiation(GO:0061141) |
| 0.5 | 2.7 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.5 | 1.6 | GO:1903006 | regulation of protein K63-linked deubiquitination(GO:1903004) positive regulation of protein K63-linked deubiquitination(GO:1903006) |
| 0.5 | 10.8 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.5 | 4.3 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.5 | 4.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.5 | 1.6 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.5 | 1.1 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.5 | 2.6 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.5 | 2.1 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.5 | 4.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.5 | 4.7 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.5 | 1.0 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.5 | 5.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.5 | 2.6 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.5 | 1.5 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.5 | 3.6 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.5 | 1.0 | GO:2000330 | positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.5 | 4.6 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.5 | 2.5 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.5 | 2.5 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.5 | 5.0 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.5 | 3.0 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.5 | 4.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.5 | 3.9 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.5 | 1.4 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.5 | 3.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.5 | 6.2 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.5 | 1.9 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.5 | 1.4 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.5 | 1.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.5 | 1.4 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.5 | 7.0 | GO:0035878 | nail development(GO:0035878) |
| 0.5 | 1.9 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.5 | 2.3 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.5 | 0.9 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.5 | 1.4 | GO:0052501 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.5 | 9.0 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.5 | 1.8 | GO:0035936 | androgen secretion(GO:0035935) testosterone secretion(GO:0035936) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.4 | 22.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.4 | 3.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.4 | 1.3 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.4 | 1.7 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.4 | 1.7 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.4 | 3.9 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.4 | 1.3 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.4 | 1.2 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.4 | 0.4 | GO:0003096 | renal sodium ion transport(GO:0003096) |
| 0.4 | 2.0 | GO:0019087 | transformation of host cell by virus(GO:0019087) renal water absorption(GO:0070295) |
| 0.4 | 7.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.4 | 1.2 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.4 | 1.6 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.4 | 1.2 | GO:0070242 | thymocyte apoptotic process(GO:0070242) |
| 0.4 | 2.4 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.4 | 5.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.4 | 1.6 | GO:0032641 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) lymphotoxin A production(GO:0032641) positive regulation of mast cell cytokine production(GO:0032765) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.4 | 1.2 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.4 | 14.0 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.4 | 5.4 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.4 | 0.8 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) positive regulation of cell maturation(GO:1903431) |
| 0.4 | 9.9 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.4 | 1.9 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.4 | 1.5 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.4 | 1.5 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.4 | 1.9 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.4 | 1.9 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.4 | 0.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.4 | 2.2 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.4 | 3.3 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.4 | 4.3 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.4 | 0.7 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.4 | 2.8 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.4 | 8.9 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.4 | 1.8 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.3 | 0.3 | GO:0035744 | T-helper 1 cell cytokine production(GO:0035744) |
| 0.3 | 2.4 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.3 | 0.7 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.3 | 1.4 | GO:0046730 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.3 | 2.4 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.3 | 3.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.3 | 3.0 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 1.3 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.3 | 1.0 | GO:0010454 | negative regulation of cell fate commitment(GO:0010454) |
| 0.3 | 0.7 | GO:0090182 | regulation of secretion of lysosomal enzymes(GO:0090182) |
| 0.3 | 1.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.3 | 1.9 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.3 | 3.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.3 | 1.0 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.3 | 3.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 0.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.3 | 2.2 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.3 | 0.9 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.3 | 2.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 1.9 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.3 | 1.5 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.3 | 3.3 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.3 | 4.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.3 | 4.8 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.3 | 1.5 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.3 | 2.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 2.7 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.3 | 1.8 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.3 | 1.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 2.6 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.3 | 2.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.3 | 1.1 | GO:1902722 | positive regulation of prolactin secretion(GO:1902722) |
| 0.3 | 4.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.3 | 9.5 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.3 | 2.2 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.3 | 1.4 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.3 | 2.7 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 1.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.3 | 1.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.3 | 3.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.3 | 8.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.3 | 3.9 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.3 | 1.6 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.3 | 3.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.3 | 0.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.3 | 0.5 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.3 | 5.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 | 1.0 | GO:0038042 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) |
| 0.3 | 1.0 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.2 | 1.7 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.2 | 2.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.2 | 4.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 1.0 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.2 | 6.0 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 6.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.2 | 1.0 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.2 | 2.1 | GO:0007135 | meiosis II(GO:0007135) |
| 0.2 | 1.6 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.2 | 1.3 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 1.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 2.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.2 | 0.9 | GO:0030047 | actin modification(GO:0030047) |
| 0.2 | 1.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.2 | 1.8 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.2 | 1.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 2.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 1.3 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 1.3 | GO:0033504 | floor plate development(GO:0033504) |
| 0.2 | 0.9 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.2 | 1.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 0.6 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.2 | 1.7 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.2 | 1.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 0.8 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 1.9 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 2.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.2 | 5.2 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.2 | 1.8 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.2 | 0.6 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.2 | 8.9 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.2 | 0.8 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.2 | 2.0 | GO:0002438 | acute inflammatory response to antigenic stimulus(GO:0002438) |
| 0.2 | 1.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 0.6 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.2 | 6.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.2 | 0.7 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.2 | 1.5 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.2 | 2.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 1.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.2 | 4.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.2 | 1.1 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 0.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.2 | 3.8 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.2 | 29.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 1.6 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 12.2 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.2 | 0.4 | GO:0051891 | regulation of cardioblast differentiation(GO:0051890) positive regulation of cardioblast differentiation(GO:0051891) |
| 0.2 | 6.2 | GO:0000732 | strand displacement(GO:0000732) |
| 0.2 | 0.9 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.2 | 1.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 0.5 | GO:0009720 | detection of hormone stimulus(GO:0009720) detection of endogenous stimulus(GO:0009726) |
| 0.2 | 4.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 1.4 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.2 | 3.9 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.2 | 0.7 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.2 | 4.8 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.2 | 1.3 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.2 | 3.4 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.2 | 2.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.2 | 0.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 0.5 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.2 | 9.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.1 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.2 | 1.4 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.2 | 0.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 0.6 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 1.4 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.2 | 1.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 6.0 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 0.6 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 | 5.9 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.2 | 1.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 1.8 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 1.6 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 3.9 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.7 | GO:0060117 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) auditory receptor cell development(GO:0060117) |
| 0.1 | 1.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 4.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 4.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.9 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.6 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 1.6 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.6 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.1 | 2.8 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.1 | 1.7 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 2.0 | GO:1904714 | chaperone-mediated autophagy(GO:0061684) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 0.4 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 2.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.5 | GO:0032909 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.1 | 0.9 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.3 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.1 | 5.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 2.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.4 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 0.7 | GO:2001206 | positive regulation of bone development(GO:1903012) positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 1.5 | GO:0090494 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.1 | 1.6 | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) |
| 0.1 | 0.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 2.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 1.3 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 3.3 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 2.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 0.6 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 3.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 1.6 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.4 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 1.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.8 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 0.3 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 1.6 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.1 | 1.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.7 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 1.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 1.3 | GO:0097152 | mesenchymal cell apoptotic process(GO:0097152) regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 3.0 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 3.7 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 0.6 | GO:0072710 | response to hydroxyurea(GO:0072710) cellular response to hydroxyurea(GO:0072711) |
| 0.1 | 1.1 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 4.7 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 1.5 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.1 | 1.5 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 1.0 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 3.7 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 4.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 1.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.7 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.7 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.1 | 0.5 | GO:0048563 | post-embryonic eye morphogenesis(GO:0048050) post-embryonic organ morphogenesis(GO:0048563) |
| 0.1 | 0.5 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 2.3 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 0.5 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.1 | 2.0 | GO:1902001 | fatty acid transmembrane transport(GO:1902001) |
| 0.1 | 2.2 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 1.3 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 0.7 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 0.6 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 0.6 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.1 | 3.0 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.1 | 2.4 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 2.1 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.1 | 2.2 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 1.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 3.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 0.4 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.6 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 6.9 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 1.4 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.9 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 2.6 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 3.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.8 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.1 | 0.3 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 3.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 2.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 7.0 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.9 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 | 1.9 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 0.5 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.1 | 0.7 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 1.0 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 2.4 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.2 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.4 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.3 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 1.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.6 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.1 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 1.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.5 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.2 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) cellular stress response to acidic pH(GO:1990451) |
| 0.1 | 0.7 | GO:0048265 | response to pain(GO:0048265) |
| 0.1 | 0.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 1.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 5.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 0.1 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.1 | 0.4 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 3.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.2 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.1 | 0.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 6.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 1.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 4.7 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 1.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 1.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.5 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 0.8 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 3.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.5 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 1.3 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 2.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 1.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 1.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 2.9 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 0.3 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 1.1 | GO:1903859 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0021612 | facial nerve structural organization(GO:0021612) |
| 0.0 | 0.5 | GO:0046621 | negative regulation of organ growth(GO:0046621) |
| 0.0 | 1.0 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 1.6 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 1.4 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0098911 | regulation of ventricular cardiac muscle cell action potential(GO:0098911) |
| 0.0 | 0.4 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 2.0 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 0.7 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.9 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) angiotensin maturation(GO:0002003) regulation of angiotensin metabolic process(GO:0060177) |
| 0.0 | 0.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.5 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.5 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.4 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.4 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.6 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.9 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 1.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 1.6 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 1.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.1 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.6 | GO:0003279 | cardiac septum development(GO:0003279) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.4 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 0.3 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 2.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.4 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.0 | 1.1 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.9 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 4.5 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.8 | GO:0015800 | acidic amino acid transport(GO:0015800) |
| 0.0 | 3.3 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 1.0 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 10.0 | GO:0007283 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 2.6 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 1.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.7 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.0 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.0 | 0.8 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) striated muscle myosin thick filament assembly(GO:0071688) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 16.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 2.9 | 8.7 | GO:0055028 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 2.5 | 10.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 2.4 | 7.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 2.2 | 6.7 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 2.0 | 9.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 1.9 | 9.6 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 1.9 | 7.4 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 1.8 | 17.9 | GO:0097443 | sorting endosome(GO:0097443) |
| 1.8 | 5.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 1.6 | 12.8 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 1.5 | 20.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.4 | 18.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.4 | 26.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 1.4 | 9.7 | GO:0043196 | varicosity(GO:0043196) |
| 1.4 | 8.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 1.3 | 3.9 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 1.3 | 8.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 1.1 | 4.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 1.1 | 8.9 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 1.0 | 4.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.9 | 14.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.9 | 11.6 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.9 | 4.5 | GO:0032449 | CBM complex(GO:0032449) |
| 0.9 | 7.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.8 | 9.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.8 | 0.8 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.8 | 4.8 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.8 | 4.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.7 | 2.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.7 | 2.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.7 | 5.0 | GO:1990745 | EARP complex(GO:1990745) |
| 0.7 | 10.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.7 | 4.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.7 | 18.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.7 | 7.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.6 | 1.9 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.6 | 1.8 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.6 | 1.8 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.6 | 8.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.6 | 6.4 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.6 | 5.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.6 | 5.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.6 | 3.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.6 | 2.2 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.5 | 1.6 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.5 | 2.6 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.5 | 2.6 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.5 | 4.7 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.5 | 1.5 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.5 | 7.0 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.5 | 2.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.5 | 4.9 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.5 | 11.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.5 | 5.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.5 | 2.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.5 | 3.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.4 | 2.7 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.4 | 12.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.4 | 3.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.4 | 8.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.4 | 2.0 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 3.7 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.4 | 6.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.4 | 2.0 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.4 | 1.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.4 | 3.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.4 | 5.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.4 | 3.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.4 | 1.8 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.4 | 7.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 2.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 4.8 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.3 | 2.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.3 | 7.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 2.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.3 | 4.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 1.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 1.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.3 | 1.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.3 | 2.0 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 3.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.3 | 4.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 1.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 17.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.3 | 0.8 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.3 | 1.6 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 1.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.3 | 0.5 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.3 | 1.3 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 1.6 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.3 | 0.8 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.3 | 5.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 1.0 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.2 | 1.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 4.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 29.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 2.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 0.7 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 3.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 3.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 1.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 5.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 0.9 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.2 | 1.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 5.9 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 4.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 1.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 1.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 1.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 1.0 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.2 | 2.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 1.2 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.2 | 1.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 3.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 3.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 1.9 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.2 | 15.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 9.8 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 2.8 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 4.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 3.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 1.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.2 | 1.9 | GO:0070449 | elongin complex(GO:0070449) |
| 0.2 | 7.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 1.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 0.5 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.2 | 6.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 9.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 1.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 8.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 5.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 1.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 0.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 3.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.9 | GO:0033180 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 5.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.8 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 10.8 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 2.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.3 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 10.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 2.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.9 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 1.0 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 1.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 1.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 3.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 3.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 3.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 4.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 8.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.5 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 1.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 5.4 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 9.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 11.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 9.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 2.0 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 1.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 0.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 8.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 5.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.3 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.6 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 5.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.6 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 7.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 17.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 1.6 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 0.6 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 6.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 3.3 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 0.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 2.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 15.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 4.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.1 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.7 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 4.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 1.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 3.5 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 2.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 7.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 1.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 26.3 | GO:0035375 | zymogen binding(GO:0035375) |
| 3.6 | 10.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 3.4 | 13.7 | GO:0032810 | sterol response element binding(GO:0032810) |
| 3.0 | 8.9 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 3.0 | 17.8 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 2.9 | 8.6 | GO:0033746 | histone demethylase activity (H3-R2 specific)(GO:0033746) histone demethylase activity (H4-R3 specific)(GO:0033749) |
| 2.8 | 8.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 2.5 | 7.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 2.5 | 15.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 2.5 | 7.5 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 2.1 | 8.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 2.0 | 6.1 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 2.0 | 18.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 2.0 | 5.9 | GO:0052815 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 1.8 | 9.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 1.8 | 17.7 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 1.7 | 5.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 1.7 | 10.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 1.7 | 6.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.6 | 12.8 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 1.6 | 6.4 | GO:0097001 | ceramide binding(GO:0097001) |
| 1.5 | 4.6 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 1.4 | 4.2 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 1.4 | 5.6 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 1.4 | 4.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 1.4 | 13.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 1.3 | 6.5 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 1.3 | 29.5 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) |
| 1.2 | 6.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 1.2 | 4.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 1.1 | 10.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 1.1 | 6.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 1.0 | 10.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 1.0 | 5.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.0 | 1.0 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 1.0 | 3.9 | GO:0050473 | arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.9 | 3.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.9 | 6.6 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.9 | 5.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.9 | 4.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.9 | 4.6 | GO:0052843 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.9 | 2.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.9 | 5.4 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.9 | 12.9 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.8 | 4.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.8 | 5.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.8 | 8.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.8 | 3.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.8 | 2.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.8 | 6.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.8 | 12.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.8 | 4.7 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.8 | 13.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.8 | 3.1 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.8 | 0.8 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.7 | 3.0 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.7 | 14.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.7 | 2.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.7 | 3.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.7 | 12.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.7 | 10.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.7 | 13.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.7 | 2.6 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.6 | 2.5 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.6 | 11.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.6 | 2.5 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.6 | 17.7 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.6 | 3.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.6 | 2.4 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.6 | 1.8 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.6 | 3.5 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.6 | 1.8 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.6 | 11.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.6 | 5.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.6 | 10.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.5 | 8.7 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.5 | 4.8 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.5 | 4.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.5 | 15.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.5 | 1.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.5 | 4.6 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.5 | 2.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.5 | 7.0 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.5 | 2.5 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.5 | 1.5 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.5 | 5.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.5 | 8.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.5 | 1.4 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.5 | 1.9 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.5 | 1.9 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.5 | 4.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.5 | 4.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.5 | 1.4 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.5 | 4.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.4 | 1.8 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.4 | 3.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.4 | 1.3 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.4 | 3.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 1.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.4 | 1.7 | GO:0004979 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.4 | 9.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.4 | 3.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.4 | 2.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.4 | 8.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.4 | 5.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.4 | 1.7 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.4 | 2.9 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.4 | 2.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.4 | 4.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.4 | 2.0 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.4 | 1.6 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.4 | 2.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.4 | 2.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.4 | 3.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.4 | 2.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.4 | 0.4 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.4 | 1.2 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.4 | 1.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.4 | 11.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.4 | 1.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 9.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.4 | 1.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.4 | 1.5 | GO:0005334 | norepinephrine:sodium symporter activity(GO:0005334) |
| 0.4 | 2.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.4 | 3.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 2.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.4 | 1.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.4 | 2.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.4 | 20.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.3 | 19.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 2.8 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 2.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.3 | 18.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 1.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.3 | 27.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.3 | 3.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 10.0 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.3 | 2.8 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.3 | 1.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.3 | 2.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.3 | 1.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.3 | 1.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.3 | 1.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.3 | 8.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 2.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 2.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 2.6 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.3 | 1.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.3 | 1.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.3 | 1.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.3 | 1.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.3 | 1.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.3 | 1.1 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.3 | 29.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 4.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.3 | 0.8 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.3 | 26.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.3 | 10.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.3 | 5.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 2.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 6.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 1.9 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.2 | 1.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 3.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 1.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 0.7 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.2 | 5.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 3.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 1.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 1.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 2.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 1.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 3.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 2.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 2.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 1.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 2.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 2.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 0.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 1.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 0.9 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 20.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 0.5 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.2 | 4.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 1.7 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 4.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 5.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 14.7 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.2 | 18.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.2 | 0.6 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.2 | 0.6 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 0.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 4.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 1.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 2.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.2 | 5.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 1.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 3.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.6 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.4 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.1 | 5.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 3.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 3.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 3.9 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 6.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 1.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 7.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 3.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.9 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 0.5 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 6.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.4 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 1.0 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 6.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 3.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 5.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 2.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 3.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.5 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 1.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 2.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 8.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 1.0 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 1.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 1.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.6 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 3.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 2.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.7 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.1 | 11.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 16.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.5 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 0.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 3.4 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.3 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 0.6 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 6.8 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 1.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 3.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.6 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 0.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 2.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 4.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 2.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.3 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.1 | 0.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.5 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.2 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 1.0 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 0.8 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 5.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 4.0 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.1 | 5.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 3.1 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 1.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 6.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.3 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 2.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 2.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 1.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 1.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.8 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 1.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 2.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.7 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 2.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 2.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 1.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 3.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 1.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 2.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 15.2 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 1.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 29.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.7 | 19.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.6 | 9.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.6 | 21.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.5 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.4 | 10.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.4 | 14.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.4 | 13.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 24.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.3 | 2.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.3 | 18.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.3 | 15.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.3 | 15.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.3 | 1.9 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 17.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 57.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 25.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.3 | 9.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.3 | 2.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.3 | 13.0 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 11.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.3 | 13.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 11.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 14.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 7.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 4.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 1.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.2 | 4.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.2 | 7.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 2.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 12.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 4.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 4.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 1.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.2 | 2.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 4.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 8.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.2 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 1.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 6.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 13.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 4.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 18.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 3.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 2.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 23.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 5.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 2.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 1.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 3.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 0.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 13.7 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.1 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 2.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 3.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 1.1 | 18.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.1 | 13.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.6 | 11.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.6 | 9.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.6 | 5.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.5 | 8.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.5 | 1.6 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.5 | 5.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.5 | 9.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.4 | 15.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.4 | 13.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.4 | 7.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.4 | 16.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.4 | 3.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 4.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.3 | 26.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.3 | 17.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 1.6 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.3 | 4.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 5.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 1.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 11.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 5.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 0.7 | REACTOME TRANSCRIPTION | Genes involved in Transcription |
| 0.2 | 9.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 16.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 2.0 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.2 | 1.6 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.2 | 11.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 4.7 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.2 | 1.7 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.2 | 5.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.2 | 3.2 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.2 | 20.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 13.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 1.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.2 | 3.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 2.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 6.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 5.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 7.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 16.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 2.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 3.5 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 3.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 5.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 2.7 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 4.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 7.2 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.1 | 4.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 4.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.9 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 1.7 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 5.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 4.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 0.6 | REACTOME SIGNALING BY GPCR | Genes involved in Signaling by GPCR |
| 0.1 | 1.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 14.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 4.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 2.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 3.2 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 1.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 8.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 9.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 1.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.7 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.1 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.1 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 2.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 4.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 0.3 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.1 | 5.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.8 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 3.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 2.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 1.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.8 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.9 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 7.5 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.5 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 1.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |