Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
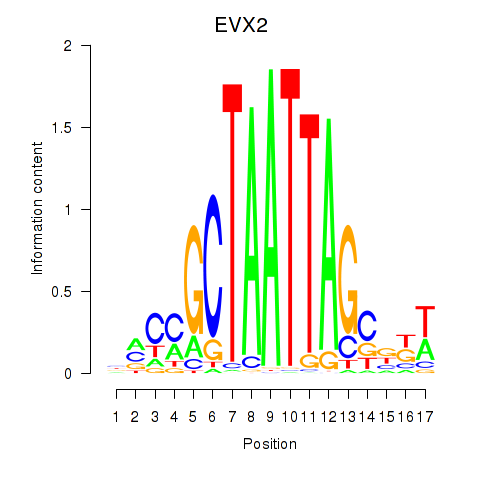
Results for EVX2
Z-value: 2.08
Transcription factors associated with EVX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EVX2
|
ENSG00000174279.4 | EVX2 |
Activity profile of EVX2 motif
Sorted Z-values of EVX2 motif
Network of associatons between targets according to the STRING database.
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_111999189 | 19.25 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chr3_+_111998739 | 19.15 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr3_+_111999326 | 18.66 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr3_+_111998915 | 18.17 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr4_+_112860981 | 16.89 |
ENST00000671704.1
|
ANK2
|
ankyrin 2 |
| chr4_+_112860912 | 16.75 |
ENST00000671951.1
|
ANK2
|
ankyrin 2 |
| chr4_+_112861053 | 16.35 |
ENST00000672221.1
|
ANK2
|
ankyrin 2 |
| chr6_+_39792298 | 14.73 |
ENST00000633794.1
ENST00000274867.9 |
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr17_-_10026265 | 9.02 |
ENST00000437099.6
ENST00000396115.6 |
GAS7
|
growth arrest specific 7 |
| chr11_-_115504389 | 8.83 |
ENST00000545380.5
ENST00000452722.7 ENST00000331581.11 ENST00000537058.5 ENST00000536727.5 ENST00000542447.6 |
CADM1
|
cell adhesion molecule 1 |
| chr21_-_31160904 | 8.17 |
ENST00000636887.1
|
TIAM1
|
TIAM Rac1 associated GEF 1 |
| chr5_+_141484997 | 8.12 |
ENST00000617094.1
ENST00000306593.2 ENST00000610539.1 ENST00000618371.4 |
PCDHGC4
|
protocadherin gamma subfamily C, 4 |
| chr1_-_151459471 | 3.91 |
ENST00000271715.7
|
POGZ
|
pogo transposable element derived with ZNF domain |
| chr6_-_62286161 | 3.87 |
ENST00000281156.5
|
KHDRBS2
|
KH RNA binding domain containing, signal transduction associated 2 |
| chr4_-_145180496 | 2.78 |
ENST00000447906.8
|
OTUD4
|
OTU deubiquitinase 4 |
| chr1_-_151459169 | 2.59 |
ENST00000368863.6
ENST00000409503.5 ENST00000491586.5 ENST00000533351.5 |
POGZ
|
pogo transposable element derived with ZNF domain |
| chr4_-_73620629 | 1.89 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6 |
| chrX_-_63755187 | 1.56 |
ENST00000635729.1
ENST00000623566.3 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9 |
| chr1_+_27935022 | 1.49 |
ENST00000411604.5
ENST00000373888.8 |
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr12_+_15546344 | 0.85 |
ENST00000674388.1
ENST00000542557.5 ENST00000445537.6 ENST00000544244.5 ENST00000442921.7 |
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr17_-_42185452 | 0.71 |
ENST00000293330.1
|
HCRT
|
hypocretin neuropeptide precursor |
| chr4_+_94974984 | 0.57 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr12_+_119668109 | 0.53 |
ENST00000229328.10
ENST00000630317.1 |
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr1_+_27934980 | 0.19 |
ENST00000373894.8
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 50.0 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.0 | 8.2 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 1.3 | 8.8 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.4 | 6.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 0.8 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 14.7 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 0.7 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.6 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 2.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 75.0 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 8.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 3.9 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 9.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 50.0 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 8.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 75.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 8.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 6.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 50.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 75.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.3 | 8.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 1.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 3.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 14.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 8.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 2.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 9.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |