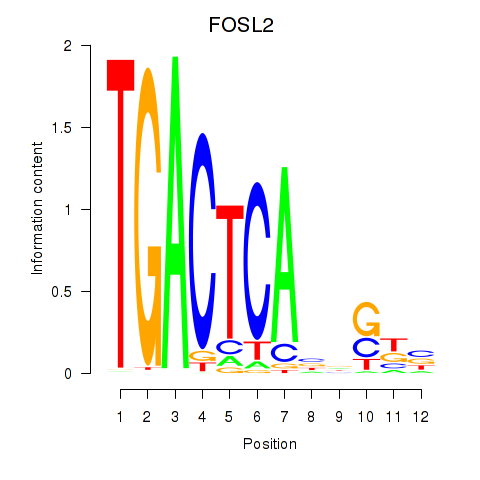
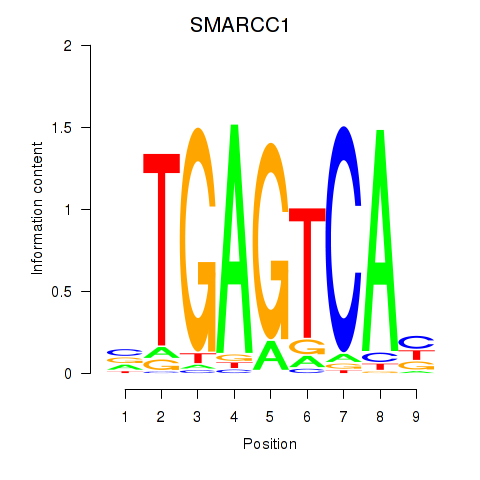
Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for FOSL2_SMARCC1
Z-value: 3.45
Transcription factors associated with FOSL2_SMARCC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSL2
|
ENSG00000075426.12 | FOSL2 |
|
SMARCC1
|
ENSG00000173473.11 | SMARCC1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMARCC1 | hg38_v1_chr3_-_47781837_47781908 | 0.34 | 3.0e-07 | Click! |
| FOSL2 | hg38_v1_chr2_+_28392802_28392866 | 0.22 | 8.3e-04 | Click! |
Activity profile of FOSL2_SMARCC1 motif
Sorted Z-values of FOSL2_SMARCC1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSL2_SMARCC1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.3 | 139.9 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 14.4 | 72.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 10.9 | 32.7 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 10.0 | 29.9 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 9.5 | 28.5 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 8.7 | 52.0 | GO:0007296 | vitellogenesis(GO:0007296) |
| 8.6 | 25.7 | GO:1902905 | positive regulation of fibril organization(GO:1902905) |
| 7.1 | 21.2 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 6.3 | 19.0 | GO:0035732 | nitric oxide storage(GO:0035732) |
| 6.0 | 29.8 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 5.2 | 73.3 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 4.9 | 49.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 4.3 | 21.4 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 4.1 | 16.5 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 3.9 | 71.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 3.7 | 11.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 3.4 | 10.2 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 3.4 | 43.8 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 3.3 | 16.5 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 3.3 | 9.9 | GO:0006113 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 3.2 | 19.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 3.2 | 9.6 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 3.2 | 9.6 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 3.2 | 15.8 | GO:1902896 | terminal web assembly(GO:1902896) |
| 3.0 | 18.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 3.0 | 14.9 | GO:0015862 | uridine transport(GO:0015862) |
| 2.8 | 8.4 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 2.7 | 19.0 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 2.7 | 40.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 2.5 | 10.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 2.3 | 6.9 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 2.2 | 6.7 | GO:0097252 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 2.2 | 20.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 2.2 | 6.7 | GO:0042946 | glucoside transport(GO:0042946) |
| 2.1 | 27.8 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 2.1 | 29.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 2.1 | 8.2 | GO:0070295 | renal water absorption(GO:0070295) |
| 2.0 | 6.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 2.0 | 5.9 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 1.9 | 28.9 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 1.7 | 17.4 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 1.7 | 5.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 1.7 | 6.8 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 1.6 | 4.8 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 1.6 | 6.2 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 1.5 | 20.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 1.4 | 12.7 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 1.4 | 11.0 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 1.2 | 24.9 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 1.2 | 29.4 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 1.2 | 4.9 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 1.2 | 6.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 1.2 | 2.4 | GO:0086098 | angiotensin-activated signaling pathway involved in heart process(GO:0086098) |
| 1.2 | 2.4 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 1.2 | 9.6 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 1.2 | 8.3 | GO:0030421 | defecation(GO:0030421) |
| 1.2 | 17.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.2 | 4.7 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 1.1 | 6.9 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 1.1 | 7.9 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 1.1 | 31.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.1 | 14.6 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 1.1 | 3.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 1.1 | 5.5 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.1 | 3.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 1.1 | 12.0 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 1.1 | 5.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 1.1 | 104.6 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 1.0 | 4.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 1.0 | 12.5 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 1.0 | 4.1 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 1.0 | 8.0 | GO:0032262 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) |
| 1.0 | 4.9 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 1.0 | 3.9 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 1.0 | 12.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 1.0 | 6.7 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.9 | 2.8 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.9 | 160.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.9 | 21.9 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.9 | 5.3 | GO:0060356 | leucine import(GO:0060356) |
| 0.9 | 13.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.9 | 12.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.8 | 3.3 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.8 | 3.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.8 | 132.6 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.8 | 42.8 | GO:0043486 | histone exchange(GO:0043486) |
| 0.8 | 21.7 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.8 | 10.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.8 | 25.6 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.8 | 5.4 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.8 | 3.8 | GO:0090270 | fibroblast growth factor production(GO:0090269) regulation of fibroblast growth factor production(GO:0090270) |
| 0.7 | 1.5 | GO:0070350 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.7 | 8.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.7 | 5.7 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.7 | 2.0 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.6 | 11.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.6 | 2.6 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.6 | 1.9 | GO:0036233 | glycine import(GO:0036233) |
| 0.6 | 3.8 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.6 | 53.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.6 | 9.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.6 | 7.5 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.6 | 8.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.6 | 4.5 | GO:0072719 | copper ion import(GO:0015677) cellular response to cisplatin(GO:0072719) |
| 0.6 | 16.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.5 | 21.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.5 | 1.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.5 | 3.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.5 | 2.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.5 | 2.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.5 | 2.3 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.5 | 5.0 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.5 | 4.1 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 6.9 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.4 | 3.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.4 | 36.8 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.4 | 9.7 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.4 | 14.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.4 | 7.5 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.4 | 3.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 1.4 | GO:0006241 | CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) |
| 0.3 | 26.7 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.3 | 7.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.3 | 12.0 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.3 | 11.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 1.0 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.3 | 15.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 1.8 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.3 | 19.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.3 | 2.9 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.3 | 2.6 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.3 | 3.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 35.9 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.3 | 1.9 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.3 | 3.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.3 | 11.8 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.3 | 1.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.3 | 1.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.3 | 11.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.3 | 25.2 | GO:0070268 | cornification(GO:0070268) |
| 0.2 | 5.7 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.2 | 1.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 0.2 | GO:0005985 | sucrose metabolic process(GO:0005985) |
| 0.2 | 0.7 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.2 | 3.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 17.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.2 | 9.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 7.2 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.2 | 0.7 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.2 | 4.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.2 | 1.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 2.3 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.2 | 2.5 | GO:1901072 | glucosamine-containing compound catabolic process(GO:1901072) |
| 0.2 | 12.3 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.2 | 1.6 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.2 | 1.0 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.2 | 2.7 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.2 | 4.8 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.2 | 0.6 | GO:0002246 | wound healing involved in inflammatory response(GO:0002246) connective tissue replacement involved in inflammatory response wound healing(GO:0002248) inflammatory response to wounding(GO:0090594) |
| 0.2 | 1.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.2 | 4.3 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.1 | 1.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 4.1 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 7.7 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 0.7 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 | 1.0 | GO:0072642 | interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.1 | 3.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 2.9 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 2.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 9.0 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 0.6 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 2.3 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 | 1.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 6.1 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 2.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 6.6 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 2.7 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.1 | 3.0 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 1.6 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 0.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 4.2 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 1.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 0.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 4.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 5.3 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 1.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 6.0 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.1 | 0.2 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 1.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.8 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 1.0 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.7 | GO:0032329 | serine transport(GO:0032329) |
| 0.1 | 3.8 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.1 | 8.0 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 2.6 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 3.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 2.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 7.1 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 7.3 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 2.4 | GO:1902402 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 | 0.3 | GO:0014010 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) Schwann cell proliferation(GO:0014010) |
| 0.0 | 2.3 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 1.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 3.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 1.2 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.8 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.7 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 0.8 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.7 | 139.9 | GO:0005638 | lamin filament(GO:0005638) |
| 10.9 | 32.7 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 9.8 | 29.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 6.1 | 73.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 5.3 | 15.8 | GO:0044393 | microspike(GO:0044393) |
| 5.1 | 25.6 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 5.0 | 9.9 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 4.8 | 19.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 4.2 | 12.5 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 4.0 | 43.8 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 4.0 | 63.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 3.4 | 20.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 3.4 | 23.8 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 3.2 | 41.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 3.2 | 28.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 3.0 | 9.1 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 2.9 | 35.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 2.9 | 20.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 2.5 | 7.5 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 2.3 | 42.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 2.1 | 19.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 2.0 | 25.7 | GO:0097413 | Lewy body(GO:0097413) |
| 1.9 | 9.6 | GO:0032449 | CBM complex(GO:0032449) |
| 1.8 | 12.5 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 1.7 | 10.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.6 | 14.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 1.6 | 70.9 | GO:0032420 | stereocilium(GO:0032420) |
| 1.6 | 51.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 1.5 | 45.5 | GO:0030057 | desmosome(GO:0030057) |
| 1.5 | 16.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.4 | 15.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 1.3 | 38.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 1.3 | 26.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 1.2 | 14.5 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 1.1 | 20.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 1.0 | 15.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 1.0 | 14.6 | GO:0097433 | dense body(GO:0097433) |
| 1.0 | 12.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.9 | 13.1 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.9 | 2.7 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.9 | 17.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.9 | 8.8 | GO:0044754 | autolysosome(GO:0044754) |
| 0.8 | 3.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.8 | 7.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.8 | 9.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.8 | 8.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.8 | 13.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.7 | 11.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.6 | 55.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.6 | 1.9 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.6 | 1.7 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.5 | 11.2 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.5 | 2.7 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.5 | 1.5 | GO:1990917 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 0.5 | 8.6 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.5 | 9.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.5 | 13.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.4 | 57.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.4 | 3.5 | GO:0030891 | VCB complex(GO:0030891) |
| 0.4 | 6.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 6.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 2.9 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.4 | 4.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.4 | 41.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.4 | 9.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 8.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 38.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.3 | 6.9 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.3 | 3.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.3 | 15.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 4.8 | GO:0031091 | platelet alpha granule(GO:0031091) platelet alpha granule lumen(GO:0031093) |
| 0.3 | 3.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 6.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.3 | 1.6 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 6.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.3 | 25.8 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 23.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 9.4 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 10.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 1.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 13.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.2 | 1.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 6.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 98.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.2 | 4.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 2.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 4.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 26.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 6.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 1.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 6.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 6.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 11.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 3.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 10.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 2.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 2.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 2.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 26.4 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 5.6 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 3.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 5.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 10.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 6.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 7.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 12.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 0.7 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 1.7 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 243.9 | GO:0044421 | extracellular region part(GO:0044421) |
| 0.1 | 1.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 14.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 3.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 4.4 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 32.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 6.3 | 19.0 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 5.5 | 16.5 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 5.5 | 32.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 4.9 | 14.6 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 4.8 | 23.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 4.1 | 20.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 3.8 | 11.4 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 3.6 | 14.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 3.4 | 10.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 3.2 | 19.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 3.0 | 21.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 2.9 | 28.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 2.5 | 14.8 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 2.4 | 97.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 2.3 | 22.6 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 2.2 | 6.7 | GO:0042947 | glucoside transmembrane transporter activity(GO:0042947) |
| 2.0 | 6.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 2.0 | 6.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 2.0 | 7.9 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 1.9 | 30.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 1.8 | 104.6 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 1.8 | 14.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.7 | 48.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 1.7 | 40.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 1.7 | 71.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 1.6 | 8.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 1.6 | 4.8 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 1.6 | 19.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 1.5 | 25.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 1.5 | 6.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 1.4 | 11.6 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 1.4 | 18.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 1.4 | 5.6 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 1.4 | 35.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 1.4 | 6.9 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 1.3 | 20.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 1.3 | 21.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.3 | 21.2 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 1.3 | 19.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 1.3 | 3.8 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 1.2 | 14.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.2 | 7.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 1.2 | 278.8 | GO:0003823 | antigen binding(GO:0003823) |
| 1.2 | 32.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 1.1 | 26.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 1.1 | 52.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 1.1 | 3.4 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 1.1 | 40.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 1.1 | 29.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 1.1 | 3.3 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 1.1 | 6.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 1.1 | 5.4 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 1.0 | 7.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 1.0 | 14.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 1.0 | 22.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 1.0 | 9.6 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.9 | 4.7 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.9 | 25.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.8 | 12.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.8 | 3.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.7 | 6.7 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.7 | 5.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.7 | 14.9 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.7 | 5.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.7 | 7.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.6 | 49.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.6 | 15.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.6 | 1.7 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.5 | 29.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.5 | 4.8 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.5 | 5.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.5 | 2.0 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.5 | 3.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.5 | 1.9 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.5 | 2.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.4 | 5.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.4 | 4.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.4 | 9.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.4 | 4.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.4 | 11.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.4 | 18.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.4 | 3.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.4 | 3.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.4 | 1.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 27.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.3 | 3.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 20.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 1.7 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.3 | 6.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.3 | 4.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 6.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 6.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.3 | 4.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 7.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 29.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 0.7 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.2 | 27.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 12.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.2 | 1.5 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 2.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 1.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 6.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 2.7 | GO:1901567 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.2 | 7.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 6.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 3.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 2.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 4.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 2.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 4.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 11.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 1.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 54.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.2 | 2.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 4.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 2.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 12.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 5.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 1.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 102.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 22.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.6 | GO:0001164 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 1.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.6 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 1.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.6 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 29.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 4.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 37.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 5.2 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 3.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 2.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.2 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.5 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 2.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.5 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 56.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 1.8 | 80.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 1.7 | 31.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.6 | 88.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 1.5 | 81.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 1.5 | 154.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 1.2 | 26.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 1.2 | 72.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 1.1 | 83.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 1.1 | 24.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 1.1 | 34.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 1.0 | 76.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 1.0 | 46.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.9 | 75.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.7 | 6.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.6 | 15.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.6 | 12.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.6 | 31.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.5 | 8.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.5 | 16.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.4 | 43.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.4 | 20.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.4 | 7.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.3 | 18.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.3 | 9.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 11.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.3 | 9.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.3 | 7.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 6.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 8.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 9.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 16.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 14.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 3.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 3.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 2.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 1.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 6.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 26.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 7.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 18.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 3.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 3.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 5.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 5.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 4.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 3.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 1.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.8 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 2.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 38.6 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 2.6 | 135.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 2.2 | 6.7 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 2.0 | 48.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 1.7 | 25.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.7 | 25.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.6 | 43.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 1.4 | 104.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 1.4 | 29.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 1.3 | 56.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 1.3 | 60.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 1.1 | 20.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 1.0 | 40.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 1.0 | 15.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.9 | 17.7 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.9 | 38.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.9 | 29.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.9 | 8.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.9 | 29.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.9 | 2.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.6 | 15.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.6 | 14.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.6 | 17.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.5 | 17.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.5 | 10.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.5 | 7.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.4 | 10.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.4 | 15.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.4 | 12.0 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.4 | 16.7 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.4 | 20.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.4 | 53.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.4 | 6.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.4 | 6.7 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.4 | 14.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.3 | 10.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.3 | 11.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.3 | 12.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.3 | 7.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.3 | 6.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.2 | 2.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 3.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 2.9 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 3.9 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 5.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 4.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.2 | 8.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 2.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 4.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 13.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 5.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 6.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 6.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 3.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 9.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 4.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 19.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 4.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 3.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 7.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 6.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 5.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |