|
chr17_-_41528293
Show fit
|
34.85 |
ENST00000455635.1
ENST00000361566.7
|
KRT19
|
keratin 19
|
|
chr5_+_136058849
Show fit
|
29.56 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced
|
|
chr18_+_3449620
Show fit
|
27.92 |
ENST00000405385.7
|
TGIF1
|
TGFB induced factor homeobox 1
|
|
chr17_-_28368012
Show fit
|
22.96 |
ENST00000555059.2
|
ENSG00000273171.1
|
novel protein, readthrough between VTN and SEBOX
|
|
chr19_+_18386150
Show fit
|
22.44 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15
|
|
chr12_-_71157872
Show fit
|
16.61 |
ENST00000546561.2
|
TSPAN8
|
tetraspanin 8
|
|
chr12_-_71157992
Show fit
|
16.05 |
ENST00000247829.8
|
TSPAN8
|
tetraspanin 8
|
|
chr12_-_89352395
Show fit
|
15.43 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr12_-_89352487
Show fit
|
14.16 |
ENST00000548755.1
ENST00000279488.8
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr12_-_52949818
Show fit
|
13.42 |
ENST00000546897.5
ENST00000552551.5
|
KRT8
|
keratin 8
|
|
chr19_-_48390847
Show fit
|
12.82 |
ENST00000597017.5
|
KDELR1
|
KDEL endoplasmic reticulum protein retention receptor 1
|
|
chr6_+_36676489
Show fit
|
12.19 |
ENST00000448526.6
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A
|
|
chr7_+_73828160
Show fit
|
11.79 |
ENST00000431918.1
|
CLDN4
|
claudin 4
|
|
chr15_+_96325935
Show fit
|
11.74 |
ENST00000421109.6
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2
|
|
chr5_+_96663010
Show fit
|
11.54 |
ENST00000506811.5
ENST00000514055.5
ENST00000508608.6
|
CAST
|
calpastatin
|
|
chr8_+_125430333
Show fit
|
11.49 |
ENST00000311922.4
|
TRIB1
|
tribbles pseudokinase 1
|
|
chr5_+_96662969
Show fit
|
10.18 |
ENST00000514845.5
ENST00000675663.1
|
CAST
|
calpastatin
|
|
chr3_+_142596385
Show fit
|
10.16 |
ENST00000457734.7
ENST00000483373.5
ENST00000475296.5
ENST00000495744.5
ENST00000476044.5
ENST00000461644.5
ENST00000464320.5
|
PLS1
|
plastin 1
|
|
chr21_-_42315336
Show fit
|
9.86 |
ENST00000398431.2
ENST00000518498.3
|
TFF3
|
trefoil factor 3
|
|
chr7_+_80602150
Show fit
|
9.67 |
ENST00000309881.11
|
CD36
|
CD36 molecule
|
|
chr7_+_80602200
Show fit
|
9.61 |
ENST00000534394.5
|
CD36
|
CD36 molecule
|
|
chr10_-_125160499
Show fit
|
9.58 |
ENST00000494626.6
ENST00000337195.9
|
CTBP2
|
C-terminal binding protein 2
|
|
chr4_-_154612635
Show fit
|
9.32 |
ENST00000407946.5
ENST00000405164.5
ENST00000336098.8
ENST00000393846.6
ENST00000404648.7
ENST00000443553.5
|
FGG
|
fibrinogen gamma chain
|
|
chrX_-_20218941
Show fit
|
9.27 |
ENST00000457145.6
|
RPS6KA3
|
ribosomal protein S6 kinase A3
|
|
chr5_+_102865805
Show fit
|
9.09 |
ENST00000346918.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr3_-_146161167
Show fit
|
8.81 |
ENST00000360060.7
ENST00000282903.10
|
PLOD2
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 2
|
|
chr7_+_134891400
Show fit
|
8.81 |
ENST00000393118.6
|
CALD1
|
caldesmon 1
|
|
chr7_+_134891566
Show fit
|
8.62 |
ENST00000424922.5
ENST00000495522.1
|
CALD1
|
caldesmon 1
|
|
chr21_-_26843012
Show fit
|
8.57 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1
|
|
chr10_+_113709261
Show fit
|
8.55 |
ENST00000672138.1
ENST00000452490.3
|
CASP7
|
caspase 7
|
|
chr10_+_102776237
Show fit
|
8.50 |
ENST00000369889.5
|
WBP1L
|
WW domain binding protein 1 like
|
|
chr3_+_186613052
Show fit
|
8.19 |
ENST00000411641.7
ENST00000273784.5
|
AHSG
|
alpha 2-HS glycoprotein
|
|
chr22_+_20774092
Show fit
|
8.18 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1
|
|
chr11_+_844406
Show fit
|
8.11 |
ENST00000397404.5
|
TSPAN4
|
tetraspanin 4
|
|
chr11_-_6440980
Show fit
|
8.08 |
ENST00000265983.8
ENST00000615166.1
|
HPX
|
hemopexin
|
|
chr21_-_26843063
Show fit
|
8.08 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1
|
|
chr10_+_122560751
Show fit
|
8.07 |
ENST00000338354.10
ENST00000664692.1
ENST00000653442.1
ENST00000664974.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr10_+_122560639
Show fit
|
8.05 |
ENST00000344338.7
ENST00000330163.8
ENST00000652446.2
ENST00000666315.1
ENST00000368955.7
ENST00000368909.7
ENST00000368956.6
ENST00000619379.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr12_-_55729660
Show fit
|
7.86 |
ENST00000546457.1
ENST00000549117.5
|
CD63
|
CD63 molecule
|
|
chr10_+_122560679
Show fit
|
7.71 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr10_-_79560386
Show fit
|
7.59 |
ENST00000372327.9
ENST00000417041.1
ENST00000640627.1
ENST00000372325.7
|
SFTPA2
|
surfactant protein A2
|
|
chr3_+_52211442
Show fit
|
7.48 |
ENST00000459884.1
|
ALAS1
|
5'-aminolevulinate synthase 1
|
|
chr11_+_62419025
Show fit
|
7.39 |
ENST00000278282.3
|
SCGB1A1
|
secretoglobin family 1A member 1
|
|
chr12_+_96194365
Show fit
|
7.32 |
ENST00000228741.8
ENST00000547249.1
|
ELK3
|
ETS transcription factor ELK3
|
|
chr12_-_91178520
Show fit
|
7.29 |
ENST00000425043.5
ENST00000420120.6
ENST00000441303.6
ENST00000456569.2
|
DCN
|
decorin
|
|
chr10_-_125161019
Show fit
|
7.27 |
ENST00000411419.6
|
CTBP2
|
C-terminal binding protein 2
|
|
chr17_-_19748355
Show fit
|
7.11 |
ENST00000494157.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1
|
|
chr19_+_16067526
Show fit
|
7.01 |
ENST00000646974.2
|
TPM4
|
tropomyosin 4
|
|
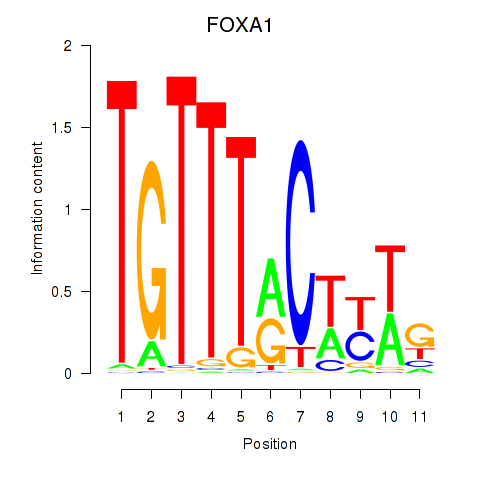
chr14_-_37595224
Show fit
|
6.93 |
ENST00000250448.5
|
FOXA1
|
forkhead box A1
|
|
chr12_-_120327762
Show fit
|
6.60 |
ENST00000308366.9
ENST00000423423.3
|
PLA2G1B
|
phospholipase A2 group IB
|
|
chr7_+_130266847
Show fit
|
6.57 |
ENST00000222481.9
|
CPA2
|
carboxypeptidase A2
|
|
chr14_+_64504743
Show fit
|
6.51 |
ENST00000683701.1
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr3_+_52777580
Show fit
|
6.31 |
ENST00000273283.7
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr2_-_21044063
Show fit
|
6.25 |
ENST00000233242.5
|
APOB
|
apolipoprotein B
|
|
chr5_-_41213505
Show fit
|
5.99 |
ENST00000337836.10
ENST00000433294.1
|
C6
|
complement C6
|
|
chr4_+_54229261
Show fit
|
5.62 |
ENST00000508170.5
ENST00000512143.1
ENST00000257290.10
|
PDGFRA
|
platelet derived growth factor receptor alpha
|
|
chr11_-_108593738
Show fit
|
5.58 |
ENST00000525344.5
ENST00000265843.9
|
EXPH5
|
exophilin 5
|
|
chr3_+_148827800
Show fit
|
5.41 |
ENST00000282957.9
ENST00000468341.1
|
CPB1
|
carboxypeptidase B1
|
|
chr10_-_46046264
Show fit
|
5.38 |
ENST00000581478.5
ENST00000582163.3
|
MSMB
|
microseminoprotein beta
|
|
chr1_-_93614091
Show fit
|
5.32 |
ENST00000370247.7
|
BCAR3
|
BCAR3 adaptor protein, NSP family member
|
|
chr6_-_107824294
Show fit
|
5.31 |
ENST00000369020.8
ENST00000369022.6
|
SCML4
|
Scm polycomb group protein like 4
|
|
chr9_-_86947496
Show fit
|
5.17 |
ENST00000298743.9
|
GAS1
|
growth arrest specific 1
|
|
chr6_+_143608170
Show fit
|
5.04 |
ENST00000427704.6
ENST00000305766.10
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr17_-_17582417
Show fit
|
4.92 |
ENST00000395783.5
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr5_+_32531786
Show fit
|
4.84 |
ENST00000512913.5
|
SUB1
|
SUB1 regulator of transcription
|
|
chr10_+_122163672
Show fit
|
4.72 |
ENST00000369004.7
ENST00000260733.7
|
TACC2
|
transforming acidic coiled-coil containing protein 2
|
|
chr5_+_180494430
Show fit
|
4.69 |
ENST00000393356.7
ENST00000618123.4
|
CNOT6
|
CCR4-NOT transcription complex subunit 6
|
|
chr15_-_55249029
Show fit
|
4.59 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr12_+_10505890
Show fit
|
4.59 |
ENST00000538173.1
|
EIF2S3B
|
eukaryotic translation initiation factor 2 subunit gamma B
|
|
chr13_+_75760659
Show fit
|
4.53 |
ENST00000526202.5
ENST00000465261.6
|
LMO7
|
LIM domain 7
|
|
chr10_+_122163590
Show fit
|
4.50 |
ENST00000368999.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2
|
|
chr1_-_94925759
Show fit
|
4.35 |
ENST00000415017.1
ENST00000545882.5
|
CNN3
|
calponin 3
|
|
chr12_-_102478539
Show fit
|
4.34 |
ENST00000424202.6
|
IGF1
|
insulin like growth factor 1
|
|
chr13_+_75760431
Show fit
|
4.21 |
ENST00000321797.12
|
LMO7
|
LIM domain 7
|
|
chr14_-_36519679
Show fit
|
4.14 |
ENST00000498187.6
|
NKX2-1
|
NK2 homeobox 1
|
|
chr4_-_69495897
Show fit
|
4.12 |
ENST00000305107.7
ENST00000639621.1
|
UGT2B4
|
UDP glucuronosyltransferase family 2 member B4
|
|
chr1_-_56966133
Show fit
|
4.06 |
ENST00000535057.5
ENST00000543257.5
|
C8B
|
complement C8 beta chain
|
|
chr21_-_37072688
Show fit
|
3.96 |
ENST00000464265.5
ENST00000399102.5
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis class P
|
|
chr4_-_69495861
Show fit
|
3.92 |
ENST00000512583.5
|
UGT2B4
|
UDP glucuronosyltransferase family 2 member B4
|
|
chr4_-_185535498
Show fit
|
3.90 |
ENST00000284767.12
ENST00000284771.7
ENST00000284770.10
ENST00000620787.5
ENST00000629667.2
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr19_-_45406327
Show fit
|
3.78 |
ENST00000593226.5
ENST00000418234.6
|
PPP1R13L
|
protein phosphatase 1 regulatory subunit 13 like
|
|
chr7_+_134745460
Show fit
|
3.76 |
ENST00000436461.6
|
CALD1
|
caldesmon 1
|
|
chrX_+_136532205
Show fit
|
3.76 |
ENST00000370634.8
|
VGLL1
|
vestigial like family member 1
|
|
chr3_+_138010143
Show fit
|
3.72 |
ENST00000183605.10
|
CLDN18
|
claudin 18
|
|
chr4_-_110198650
Show fit
|
3.65 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6
|
|
chr12_+_120978686
Show fit
|
3.64 |
ENST00000541395.5
ENST00000544413.2
|
HNF1A
|
HNF1 homeobox A
|
|
chr12_+_21372899
Show fit
|
3.64 |
ENST00000240652.8
ENST00000542023.1
ENST00000537593.1
|
IAPP
|
islet amyloid polypeptide
|
|
chr6_-_87095059
Show fit
|
3.58 |
ENST00000369582.6
ENST00000610310.3
ENST00000630630.2
ENST00000627148.3
ENST00000625577.1
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr14_-_94323324
Show fit
|
3.58 |
ENST00000341584.4
|
SERPINA6
|
serpin family A member 6
|
|
chr1_-_56966006
Show fit
|
3.55 |
ENST00000371237.9
|
C8B
|
complement C8 beta chain
|
|
chr14_-_36582593
Show fit
|
3.48 |
ENST00000258829.6
|
NKX2-8
|
NK2 homeobox 8
|
|
chr17_-_47957824
Show fit
|
3.46 |
ENST00000300557.3
|
PRR15L
|
proline rich 15 like
|
|
chr11_-_102530738
Show fit
|
3.42 |
ENST00000260227.5
|
MMP7
|
matrix metallopeptidase 7
|
|
chr7_+_95485898
Show fit
|
3.40 |
ENST00000428113.5
|
ASB4
|
ankyrin repeat and SOCS box containing 4
|
|
chr7_-_100100716
Show fit
|
3.38 |
ENST00000354230.7
ENST00000425308.5
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr5_+_180494344
Show fit
|
3.33 |
ENST00000261951.9
|
CNOT6
|
CCR4-NOT transcription complex subunit 6
|
|
chr20_+_33235987
Show fit
|
3.21 |
ENST00000375422.6
ENST00000375413.8
ENST00000354297.9
|
BPIFA1
|
BPI fold containing family A member 1
|
|
chr2_+_168901290
Show fit
|
3.13 |
ENST00000429379.2
ENST00000375363.8
ENST00000421979.1
|
G6PC2
|
glucose-6-phosphatase catalytic subunit 2
|
|
chr3_-_114179052
Show fit
|
3.04 |
ENST00000383673.5
ENST00000295881.9
|
DRD3
|
dopamine receptor D3
|
|
chr3_+_137998735
Show fit
|
2.97 |
ENST00000343735.8
|
CLDN18
|
claudin 18
|
|
chr7_+_112423137
Show fit
|
2.93 |
ENST00000005558.8
ENST00000621379.4
|
IFRD1
|
interferon related developmental regulator 1
|
|
chr1_-_244860376
Show fit
|
2.88 |
ENST00000638716.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U
|
|
chr4_+_146175702
Show fit
|
2.77 |
ENST00000296581.11
ENST00000649747.1
ENST00000502781.5
|
LSM6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated
|
|
chr18_+_68798065
Show fit
|
2.65 |
ENST00000360242.9
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr1_+_200027605
Show fit
|
2.64 |
ENST00000236914.7
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2
|
|
chr5_-_138338325
Show fit
|
2.61 |
ENST00000510119.1
ENST00000513970.5
|
CDC25C
|
cell division cycle 25C
|
|
chr10_-_79949098
Show fit
|
2.53 |
ENST00000372292.8
|
SFTPD
|
surfactant protein D
|
|
chr6_-_41747390
Show fit
|
2.52 |
ENST00000356667.8
ENST00000373025.7
ENST00000425343.6
|
PGC
|
progastricsin
|
|
chr8_+_32646838
Show fit
|
2.52 |
ENST00000651333.1
ENST00000652592.1
|
NRG1
|
neuregulin 1
|
|
chr1_-_203351115
Show fit
|
2.50 |
ENST00000354955.5
|
FMOD
|
fibromodulin
|
|
chr8_+_32647080
Show fit
|
2.49 |
ENST00000520502.7
ENST00000523041.2
ENST00000650819.1
|
NRG1
|
neuregulin 1
|
|
chr3_+_141387801
Show fit
|
2.48 |
ENST00000514251.5
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr11_-_10568650
Show fit
|
2.46 |
ENST00000256178.8
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1
|
|
chr20_-_7940444
Show fit
|
2.46 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1
|
|
chr12_-_48004467
Show fit
|
2.45 |
ENST00000380518.8
|
COL2A1
|
collagen type II alpha 1 chain
|
|
chr7_+_28412511
Show fit
|
2.42 |
ENST00000357727.7
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr9_-_92424427
Show fit
|
2.41 |
ENST00000375550.5
|
OMD
|
osteomodulin
|
|
chr8_+_69492793
Show fit
|
2.29 |
ENST00000616868.1
ENST00000419716.7
ENST00000402687.9
|
SULF1
|
sulfatase 1
|
|
chr11_+_121102666
Show fit
|
2.29 |
ENST00000264037.2
|
TECTA
|
tectorin alpha
|
|
chr22_-_21227637
Show fit
|
2.27 |
ENST00000401924.5
|
GGT2
|
gamma-glutamyltransferase 2
|
|
chr7_+_77840122
Show fit
|
2.27 |
ENST00000450574.5
ENST00000248550.7
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr17_+_9021501
Show fit
|
2.20 |
ENST00000173229.7
|
NTN1
|
netrin 1
|
|
chr12_-_48004496
Show fit
|
2.17 |
ENST00000337299.7
|
COL2A1
|
collagen type II alpha 1 chain
|
|
chr6_+_160702238
Show fit
|
2.14 |
ENST00000366924.6
ENST00000308192.14
ENST00000418964.1
|
PLG
|
plasminogen
|
|
chr13_+_77535669
Show fit
|
2.09 |
ENST00000535157.5
|
SCEL
|
sciellin
|
|
chr13_+_77535681
Show fit
|
2.09 |
ENST00000349847.4
|
SCEL
|
sciellin
|
|
chr17_+_76376581
Show fit
|
2.08 |
ENST00000591651.5
ENST00000545180.5
|
SPHK1
|
sphingosine kinase 1
|
|
chr10_+_101131284
Show fit
|
2.07 |
ENST00000370196.11
ENST00000467928.2
|
TLX1
|
T cell leukemia homeobox 1
|
|
chr1_-_116667668
Show fit
|
2.07 |
ENST00000369486.8
ENST00000369483.5
|
IGSF3
|
immunoglobulin superfamily member 3
|
|
chr13_+_77535742
Show fit
|
2.00 |
ENST00000377246.7
|
SCEL
|
sciellin
|
|
chr9_+_27109200
Show fit
|
1.98 |
ENST00000380036.10
|
TEK
|
TEK receptor tyrosine kinase
|
|
chr11_+_59787067
Show fit
|
1.92 |
ENST00000528805.1
|
STX3
|
syntaxin 3
|
|
chr1_-_216423396
Show fit
|
1.89 |
ENST00000366942.3
ENST00000674083.1
ENST00000307340.8
|
USH2A
|
usherin
|
|
chr16_-_48610150
Show fit
|
1.88 |
ENST00000262384.4
|
N4BP1
|
NEDD4 binding protein 1
|
|
chr4_-_110198579
Show fit
|
1.87 |
ENST00000302274.8
|
ELOVL6
|
ELOVL fatty acid elongase 6
|
|
chr3_+_69936583
Show fit
|
1.84 |
ENST00000314557.10
ENST00000394351.9
|
MITF
|
melanocyte inducing transcription factor
|
|
chr15_-_29822418
Show fit
|
1.82 |
ENST00000614355.5
ENST00000495972.6
ENST00000346128.10
|
TJP1
|
tight junction protein 1
|
|
chrX_+_100584928
Show fit
|
1.82 |
ENST00000373031.5
|
TNMD
|
tenomodulin
|
|
chr1_+_74235377
Show fit
|
1.80 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr9_+_27109135
Show fit
|
1.75 |
ENST00000519097.5
ENST00000615002.4
|
TEK
|
TEK receptor tyrosine kinase
|
|
chr7_+_48924559
Show fit
|
1.72 |
ENST00000650262.1
|
CDC14C
|
cell division cycle 14C
|
|
chr1_-_246566238
Show fit
|
1.70 |
ENST00000366514.5
|
TFB2M
|
transcription factor B2, mitochondrial
|
|
chr5_-_136365476
Show fit
|
1.68 |
ENST00000378459.7
ENST00000502753.4
ENST00000513104.6
ENST00000352189.8
|
TRPC7
|
transient receptor potential cation channel subfamily C member 7
|
|
chr2_+_28395511
Show fit
|
1.63 |
ENST00000436647.1
|
FOSL2
|
FOS like 2, AP-1 transcription factor subunit
|
|
chr6_-_42048648
Show fit
|
1.61 |
ENST00000502771.1
ENST00000508143.5
ENST00000514588.1
ENST00000510503.5
|
CCND3
|
cyclin D3
|
|
chr7_+_135148041
Show fit
|
1.59 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140
|
|
chr5_+_96936071
Show fit
|
1.55 |
ENST00000231368.10
|
LNPEP
|
leucyl and cystinyl aminopeptidase
|
|
chr11_+_1157946
Show fit
|
1.53 |
ENST00000621226.2
|
MUC5AC
|
mucin 5AC, oligomeric mucus/gel-forming
|
|
chr1_+_152514474
Show fit
|
1.51 |
ENST00000368790.4
|
CRCT1
|
cysteine rich C-terminal 1
|
|
chr14_+_64503943
Show fit
|
1.50 |
ENST00000556965.1
ENST00000554015.5
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr1_+_200027702
Show fit
|
1.49 |
ENST00000367362.8
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2
|
|
chr1_+_116111395
Show fit
|
1.48 |
ENST00000684484.1
ENST00000369500.4
|
MAB21L3
|
mab-21 like 3
|
|
chr5_+_148394712
Show fit
|
1.46 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38
|
|
chr7_-_81770039
Show fit
|
1.43 |
ENST00000222390.11
ENST00000453411.6
ENST00000457544.7
ENST00000444829.7
|
HGF
|
hepatocyte growth factor
|
|
chr9_+_79572572
Show fit
|
1.36 |
ENST00000435650.5
ENST00000414465.5
ENST00000376537.8
|
TLE4
|
TLE family member 4, transcriptional corepressor
|
|
chr21_-_14658812
Show fit
|
1.36 |
ENST00000647101.1
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr12_-_14696571
Show fit
|
1.30 |
ENST00000261170.5
|
GUCY2C
|
guanylate cyclase 2C
|
|
chrX_-_15384402
Show fit
|
1.26 |
ENST00000297904.4
|
VEGFD
|
vascular endothelial growth factor D
|
|
chr2_+_151409878
Show fit
|
1.24 |
ENST00000453091.6
ENST00000428287.6
ENST00000444746.7
ENST00000243326.9
ENST00000414861.6
|
RIF1
|
replication timing regulatory factor 1
|
|
chr7_+_117014881
Show fit
|
1.16 |
ENST00000422922.5
ENST00000432298.5
|
ST7
|
suppression of tumorigenicity 7
|
|
chr13_-_48413105
Show fit
|
1.15 |
ENST00000620633.5
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr4_+_186227501
Show fit
|
1.14 |
ENST00000446598.6
ENST00000264690.11
ENST00000513864.2
|
KLKB1
|
kallikrein B1
|
|
chr2_-_171160833
Show fit
|
1.14 |
ENST00000360843.7
ENST00000431350.7
|
TLK1
|
tousled like kinase 1
|
|
chr3_+_148730100
Show fit
|
1.12 |
ENST00000474935.5
ENST00000475347.5
ENST00000461609.1
|
AGTR1
|
angiotensin II receptor type 1
|
|
chr20_+_64164566
Show fit
|
1.11 |
ENST00000650655.1
|
MYT1
|
myelin transcription factor 1
|
|
chr12_+_120978537
Show fit
|
1.06 |
ENST00000257555.11
ENST00000400024.6
|
HNF1A
|
HNF1 homeobox A
|
|
chr12_+_103587266
Show fit
|
1.04 |
ENST00000388887.7
|
STAB2
|
stabilin 2
|
|
chr8_+_132866948
Show fit
|
1.04 |
ENST00000220616.9
|
TG
|
thyroglobulin
|
|
chr8_+_11809135
Show fit
|
1.03 |
ENST00000528643.5
ENST00000525777.5
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1
|
|
chr20_+_64164446
Show fit
|
0.99 |
ENST00000328439.6
|
MYT1
|
myelin transcription factor 1
|
|
chr6_+_52420992
Show fit
|
0.99 |
ENST00000636954.1
ENST00000636566.1
ENST00000638075.1
|
EFHC1
|
EF-hand domain containing 1
|
|
chr8_+_76681208
Show fit
|
0.93 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr2_-_213151590
Show fit
|
0.92 |
ENST00000374319.8
ENST00000457361.5
ENST00000451136.6
ENST00000434687.6
|
IKZF2
|
IKAROS family zinc finger 2
|
|
chr2_-_49154507
Show fit
|
0.87 |
ENST00000406846.7
|
FSHR
|
follicle stimulating hormone receptor
|
|
chr20_+_64164474
Show fit
|
0.86 |
ENST00000622439.4
ENST00000536311.5
|
MYT1
|
myelin transcription factor 1
|
|
chr9_+_79572715
Show fit
|
0.84 |
ENST00000265284.10
|
TLE4
|
TLE family member 4, transcriptional corepressor
|
|
chr17_+_69414690
Show fit
|
0.81 |
ENST00000590474.7
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr5_-_128339191
Show fit
|
0.81 |
ENST00000507835.5
|
FBN2
|
fibrillin 2
|
|
chr6_-_160664270
Show fit
|
0.72 |
ENST00000316300.10
|
LPA
|
lipoprotein(a)
|
|
chrX_-_24647091
Show fit
|
0.72 |
ENST00000356768.8
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr2_+_47403061
Show fit
|
0.71 |
ENST00000543555.6
|
MSH2
|
mutS homolog 2
|
|
chrX_+_136648138
Show fit
|
0.70 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand
|
|
chr14_+_36661852
Show fit
|
0.70 |
ENST00000361487.7
|
PAX9
|
paired box 9
|
|
chr10_+_116590956
Show fit
|
0.70 |
ENST00000358834.9
ENST00000528052.5
|
PNLIPRP1
|
pancreatic lipase related protein 1
|
|
chr3_+_160677152
Show fit
|
0.67 |
ENST00000320767.4
|
ARL14
|
ADP ribosylation factor like GTPase 14
|
|
chr3_+_189631373
Show fit
|
0.67 |
ENST00000264731.8
ENST00000418709.6
ENST00000320472.9
ENST00000392460.7
ENST00000440651.6
|
TP63
|
tumor protein p63
|
|
chr3_+_69936629
Show fit
|
0.67 |
ENST00000394348.2
ENST00000531774.1
|
MITF
|
melanocyte inducing transcription factor
|
|
chr2_-_49154433
Show fit
|
0.65 |
ENST00000454032.5
ENST00000304421.8
|
FSHR
|
follicle stimulating hormone receptor
|
|
chr6_-_89217339
Show fit
|
0.60 |
ENST00000454853.7
|
GABRR1
|
gamma-aminobutyric acid type A receptor subunit rho1
|
|
chr14_-_64504570
Show fit
|
0.59 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25
|
|
chr7_+_106865263
Show fit
|
0.56 |
ENST00000440650.6
ENST00000496166.6
ENST00000473541.5
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma
|
|
chr19_-_18791297
Show fit
|
0.55 |
ENST00000542601.6
ENST00000222271.7
ENST00000425807.1
|
COMP
|
cartilage oligomeric matrix protein
|
|
chrX_+_1268807
Show fit
|
0.55 |
ENST00000381524.8
ENST00000381529.9
ENST00000412290.6
|
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha
|
|
chr2_+_203936755
Show fit
|
0.53 |
ENST00000316386.11
ENST00000435193.1
|
ICOS
|
inducible T cell costimulator
|
|
chr12_-_52434363
Show fit
|
0.53 |
ENST00000252245.6
|
KRT75
|
keratin 75
|
|
chr9_-_20622479
Show fit
|
0.51 |
ENST00000380338.9
|
MLLT3
|
MLLT3 super elongation complex subunit
|
|
chr17_-_50468871
Show fit
|
0.51 |
ENST00000508540.6
ENST00000258969.4
|
CHAD
|
chondroadherin
|
|
chr2_+_70935919
Show fit
|
0.44 |
ENST00000412314.5
|
ATP6V1B1
|
ATPase H+ transporting V1 subunit B1
|
|
chrX_+_16946862
Show fit
|
0.43 |
ENST00000303843.7
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr7_+_30963945
Show fit
|
0.43 |
ENST00000326139.7
|
GHRHR
|
growth hormone releasing hormone receptor
|