Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
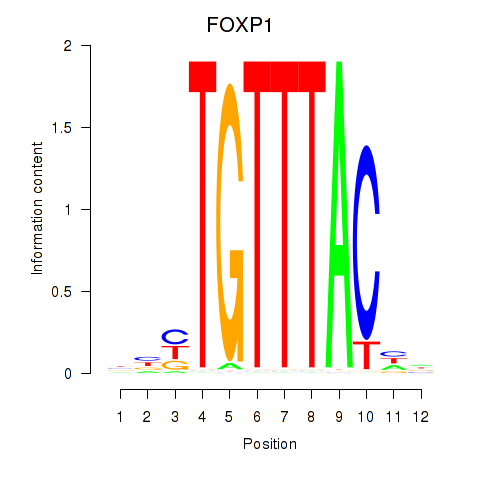
Results for FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
Z-value: 7.54
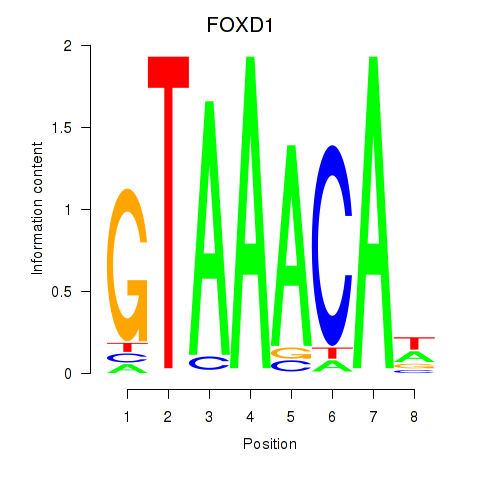
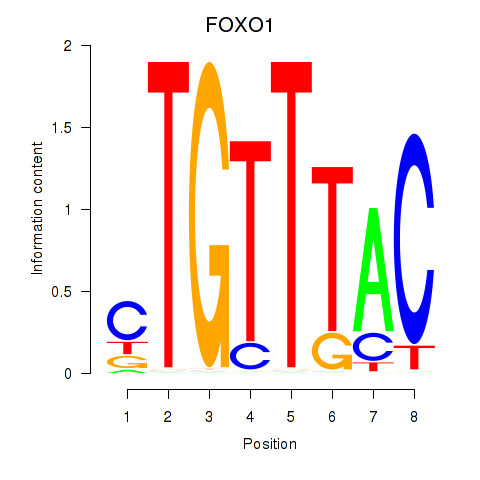
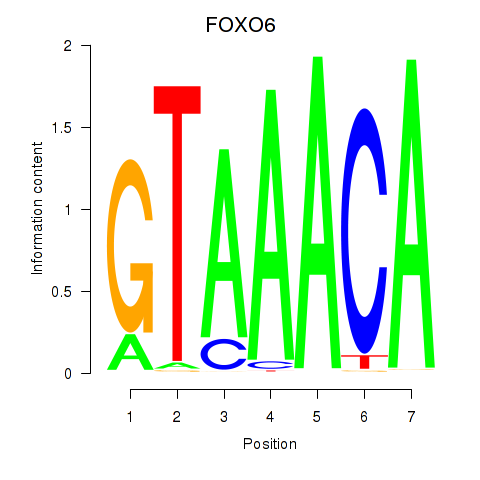
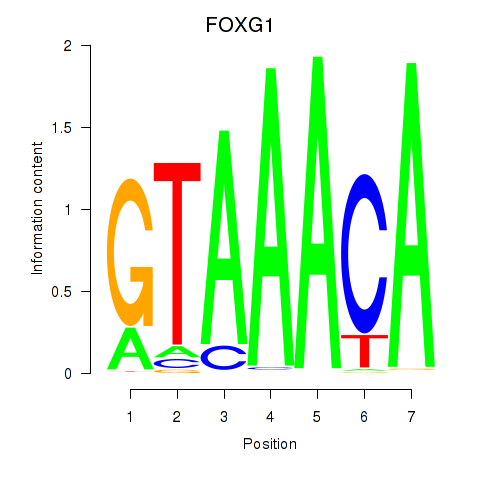
Transcription factors associated with FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXD1
|
ENSG00000251493.5 | FOXD1 |
|
FOXO1
|
ENSG00000150907.10 | FOXO1 |
|
FOXO6
|
ENSG00000204060.7 | FOXO6 |
|
FOXG1
|
ENSG00000176165.12 | FOXG1 |
|
FOXP1
|
ENSG00000114861.23 | FOXP1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXO1 | hg38_v1_chr13_-_40666600_40666650 | 0.59 | 3.1e-22 | Click! |
| FOXD1 | hg38_v1_chr5_-_73448769_73448784 | -0.36 | 5.7e-08 | Click! |
| FOXG1 | hg38_v1_chr14_+_28766755_28766802 | -0.05 | 5.0e-01 | Click! |
Activity profile of FOXD1_FOXO1_FOXO6_FOXG1_FOXP1 motif
Sorted Z-values of FOXD1_FOXO1_FOXO6_FOXG1_FOXP1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.2 | 112.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 7.7 | 23.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 7.0 | 28.1 | GO:1902617 | response to fluoride(GO:1902617) |
| 6.6 | 46.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 6.4 | 32.2 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 6.0 | 18.0 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 5.4 | 42.9 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 4.3 | 55.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 4.1 | 16.5 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 4.0 | 11.9 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 3.7 | 29.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 3.6 | 32.6 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 3.6 | 14.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 3.5 | 10.6 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 3.5 | 21.0 | GO:0097338 | response to clozapine(GO:0097338) |
| 3.3 | 10.0 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 3.3 | 32.6 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 3.1 | 12.6 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 3.1 | 18.6 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 3.1 | 40.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 2.8 | 8.5 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 2.8 | 25.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 2.7 | 19.0 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 2.4 | 17.0 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 2.4 | 11.8 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 2.3 | 7.0 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 2.3 | 20.9 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 2.3 | 6.9 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 2.2 | 10.9 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 2.1 | 10.7 | GO:1990164 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 2.1 | 10.3 | GO:0051029 | rRNA transport(GO:0051029) |
| 2.0 | 6.1 | GO:0045957 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 2.0 | 16.0 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 2.0 | 15.6 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 1.9 | 39.0 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 1.9 | 57.1 | GO:0002347 | response to tumor cell(GO:0002347) |
| 1.9 | 16.9 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 1.9 | 5.6 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 1.9 | 5.6 | GO:0042704 | uterine wall breakdown(GO:0042704) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 1.8 | 30.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 1.7 | 7.0 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 1.7 | 6.7 | GO:1990737 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 1.7 | 28.1 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.6 | 12.8 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 1.6 | 6.4 | GO:0010157 | response to chlorate(GO:0010157) |
| 1.5 | 3.0 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 1.5 | 4.4 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 1.4 | 2.9 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 1.4 | 8.3 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 1.3 | 1.3 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 1.3 | 8.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 1.3 | 22.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.3 | 6.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 1.2 | 5.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 1.2 | 6.0 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 1.1 | 8.7 | GO:0009597 | detection of virus(GO:0009597) |
| 1.1 | 4.3 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 1.1 | 3.2 | GO:0051136 | regulation of extrathymic T cell differentiation(GO:0033082) regulation of NK T cell differentiation(GO:0051136) sebum secreting cell proliferation(GO:1990654) |
| 1.1 | 15.1 | GO:0098909 | maintenance of centrosome location(GO:0051661) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 1.1 | 3.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 1.1 | 4.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 1.0 | 8.4 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 1.0 | 3.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.0 | 42.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 1.0 | 6.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 1.0 | 3.0 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 1.0 | 12.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.0 | 6.0 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 1.0 | 6.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.0 | 22.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.9 | 1.8 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.9 | 4.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.8 | 5.8 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.8 | 15.6 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.8 | 9.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.8 | 19.5 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.8 | 12.2 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.8 | 2.4 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.8 | 15.2 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.8 | 2.4 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.8 | 3.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.8 | 3.9 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.8 | 7.7 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.8 | 5.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.8 | 7.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.7 | 2.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.7 | 3.7 | GO:0044860 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) protein localization to plasma membrane raft(GO:0044860) |
| 0.7 | 12.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.7 | 3.7 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.7 | 3.7 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.7 | 5.8 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.7 | 21.5 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.7 | 7.3 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.7 | 3.3 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.7 | 3.9 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.6 | 48.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.6 | 4.4 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.6 | 2.5 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.6 | 1.8 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.6 | 0.6 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.6 | 2.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.6 | 1.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.6 | 3.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.5 | 3.2 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.5 | 3.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.5 | 2.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.5 | 2.0 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.5 | 1.5 | GO:0060366 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.5 | 2.5 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.5 | 3.5 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.5 | 1.0 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.5 | 0.5 | GO:1902227 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.5 | 1.9 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.5 | 1.4 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.5 | 5.7 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.5 | 1.8 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar Purkinje cell layer maturation(GO:0021691) cerebellar cortex maturation(GO:0021699) |
| 0.5 | 0.9 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.4 | 11.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.4 | 3.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.4 | 13.2 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.4 | 4.7 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.4 | 1.3 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.4 | 3.8 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.4 | 7.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.4 | 109.0 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.4 | 2.9 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.4 | 4.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.4 | 3.2 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.4 | 6.0 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.4 | 7.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.4 | 12.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.4 | 3.0 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.4 | 4.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.4 | 7.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.4 | 1.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.4 | 5.9 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) ATF6-mediated unfolded protein response(GO:0036500) |
| 0.4 | 1.4 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.4 | 1.4 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.4 | 2.1 | GO:0060594 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) mammary gland specification(GO:0060594) |
| 0.3 | 2.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.3 | 4.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.3 | 10.9 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.3 | 1.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.3 | 1.3 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.3 | 2.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.3 | 5.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.3 | 25.0 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.3 | 0.9 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.3 | 5.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 8.7 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.3 | 24.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.3 | 3.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.3 | 3.5 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.3 | 9.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.3 | 1.9 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 2.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 2.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 2.2 | GO:0015825 | L-serine transport(GO:0015825) serine transport(GO:0032329) |
| 0.2 | 0.5 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 | 2.9 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 1.7 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.2 | 4.0 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.2 | 4.7 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.2 | 1.2 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.2 | 1.8 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.2 | 9.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 0.7 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.2 | 13.7 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.2 | 18.8 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.2 | 0.7 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.2 | 1.5 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.2 | 1.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.8 | GO:0010193 | response to ozone(GO:0010193) |
| 0.2 | 12.0 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.2 | 0.8 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 0.2 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.2 | 1.6 | GO:0090166 | vesicle fusion with Golgi apparatus(GO:0048280) Golgi disassembly(GO:0090166) |
| 0.2 | 4.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 1.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 1.2 | GO:0001999 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) |
| 0.2 | 0.6 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 1.9 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.2 | 4.3 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.2 | 1.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 0.7 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.2 | 2.1 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 1.8 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 4.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 1.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 1.7 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.2 | 6.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 2.7 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.2 | 0.5 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.2 | 3.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.2 | 10.3 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.2 | 0.6 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.2 | 2.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 7.9 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 0.6 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 2.2 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.2 | 0.9 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.1 | 1.8 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 22.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 1.4 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 2.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 3.8 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 2.4 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 0.7 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 1.0 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.4 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 7.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 1.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 9.5 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.1 | 0.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.2 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) negative regulation of gastrulation(GO:2000542) |
| 0.1 | 0.4 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.1 | 4.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.8 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 2.2 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.1 | 1.9 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.1 | 3.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 1.2 | GO:0046851 | negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.1 | 0.9 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.1 | 1.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 2.2 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.1 | 1.6 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 1.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 4.3 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 0.4 | GO:0001519 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.1 | 0.7 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.1 | 0.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.7 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 0.4 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 2.1 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 1.6 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.5 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.2 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 0.1 | 0.7 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.4 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.1 | 1.3 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.1 | 1.9 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 0.4 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 5.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 8.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.9 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 0.4 | GO:1900368 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.3 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 0.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.4 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.1 | 0.8 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.7 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.1 | 3.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 3.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.2 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.3 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.3 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 1.9 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.1 | 1.1 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 0.2 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 1.1 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.1 | 0.1 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) melanocyte proliferation(GO:0097325) |
| 0.0 | 0.7 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 1.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.6 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.5 | GO:0051195 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 0.2 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.0 | 3.7 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 2.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.4 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.3 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 15.6 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.8 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 1.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 1.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.8 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 1.1 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:0044321 | response to leptin(GO:0044321) |
| 0.0 | 0.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 1.1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 1.9 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.3 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.2 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.5 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.3 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 1.6 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.6 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.0 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.6 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 2.1 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.0 | 0.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 35.9 | GO:0036398 | TCR signalosome(GO:0036398) |
| 5.6 | 16.7 | GO:0044609 | DBIRD complex(GO:0044609) |
| 4.8 | 38.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 3.8 | 15.1 | GO:0044307 | dendritic branch(GO:0044307) |
| 3.3 | 9.8 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 2.6 | 10.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 1.9 | 17.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.8 | 5.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 1.7 | 16.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 1.5 | 39.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 1.4 | 7.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 1.4 | 27.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 1.3 | 21.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.3 | 3.9 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.3 | 30.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 1.2 | 40.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 1.1 | 5.7 | GO:0035838 | growing cell tip(GO:0035838) |
| 1.1 | 4.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 1.1 | 12.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 1.0 | 3.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.0 | 18.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 1.0 | 21.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.9 | 12.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.9 | 37.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.8 | 5.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.7 | 6.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.6 | 7.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.6 | 1.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.6 | 50.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.6 | 2.9 | GO:0005712 | chiasma(GO:0005712) |
| 0.5 | 4.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.5 | 2.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.5 | 103.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.5 | 6.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.5 | 1.5 | GO:0034657 | GID complex(GO:0034657) |
| 0.5 | 4.3 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.4 | 5.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 11.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.4 | 3.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.4 | 2.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.4 | 1.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.3 | 2.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.3 | 3.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 3.7 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.3 | 3.4 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.3 | 4.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.3 | 138.8 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.3 | 3.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 2.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.3 | 2.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 1.3 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.3 | 2.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 10.5 | GO:0016514 | SWI/SNF complex(GO:0016514) npBAF complex(GO:0071564) |
| 0.3 | 0.9 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 13.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 2.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 2.9 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 3.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 5.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 8.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 1.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 3.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 3.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 5.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 1.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 0.7 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.2 | 1.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 1.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 6.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 19.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.4 | GO:0070701 | mucus layer(GO:0070701) |
| 0.1 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 16.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 13.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 26.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 7.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 7.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 11.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 2.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 7.6 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 1.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 18.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 8.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 3.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 5.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 9.8 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 2.4 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.1 | 6.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 1.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 31.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 18.0 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 3.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.4 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 18.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 12.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 3.6 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 2.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 6.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 53.5 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 9.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.7 | 112.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 8.1 | 32.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 5.8 | 121.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 5.0 | 29.7 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 4.6 | 41.7 | GO:0043426 | MRF binding(GO:0043426) |
| 3.7 | 18.7 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 3.5 | 10.6 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 3.3 | 10.0 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 3.3 | 46.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 2.8 | 8.5 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 2.7 | 18.6 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 2.5 | 9.8 | GO:0002046 | opsin binding(GO:0002046) |
| 2.5 | 29.4 | GO:0008430 | selenium binding(GO:0008430) |
| 2.0 | 6.1 | GO:0004877 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 2.0 | 12.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.9 | 7.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.9 | 5.7 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 1.9 | 16.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 1.8 | 5.5 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 1.7 | 5.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 1.7 | 5.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 1.6 | 6.4 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 1.5 | 18.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 1.5 | 5.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.4 | 4.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.3 | 14.8 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 1.3 | 6.7 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 1.3 | 21.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 1.3 | 25.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 1.2 | 5.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 1.2 | 28.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 1.2 | 4.7 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 1.2 | 14.0 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 1.2 | 27.9 | GO:0030275 | LRR domain binding(GO:0030275) |
| 1.1 | 3.4 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 1.1 | 5.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.1 | 22.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 1.1 | 4.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 1.0 | 8.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 1.0 | 35.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 1.0 | 10.9 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 1.0 | 16.5 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 1.0 | 3.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.9 | 22.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.9 | 4.5 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.9 | 14.4 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.9 | 1.8 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.9 | 7.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.9 | 12.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.8 | 9.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.8 | 4.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.7 | 10.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.7 | 2.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.7 | 21.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.7 | 6.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.7 | 31.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.7 | 13.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.6 | 17.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.6 | 1.8 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.6 | 15.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.6 | 1.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.6 | 2.9 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.6 | 1.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.6 | 3.4 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.6 | 3.9 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.5 | 3.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.5 | 2.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.5 | 5.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.5 | 12.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.5 | 2.9 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.4 | 5.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 1.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 6.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.4 | 11.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.4 | 2.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.4 | 5.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.4 | 7.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 10.0 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.4 | 10.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.4 | 2.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.4 | 4.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.4 | 1.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.4 | 2.9 | GO:0005534 | galactose binding(GO:0005534) |
| 0.4 | 16.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.3 | 1.7 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.3 | 2.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 4.4 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.3 | 5.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 1.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.3 | 27.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 2.5 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.3 | 1.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 1.8 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.3 | 1.8 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 2.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 53.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.3 | 0.9 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.3 | 20.7 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.3 | 3.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 1.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 2.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 5.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 16.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 5.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 0.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.3 | 9.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 4.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 5.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 9.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 7.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.3 | 1.0 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.2 | 0.7 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.2 | 6.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 0.7 | GO:0004577 | N-acetylglucosaminyldiphosphodolichol N-acetylglucosaminyltransferase activity(GO:0004577) |
| 0.2 | 3.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 63.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.2 | 0.7 | GO:0031711 | angiotensin type I receptor activity(GO:0001596) bradykinin receptor binding(GO:0031711) |
| 0.2 | 2.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 1.9 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 1.6 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.2 | 11.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 3.5 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.2 | 1.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 1.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 0.7 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.2 | 11.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 2.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 4.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 3.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 3.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 3.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 0.5 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.2 | 0.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 0.6 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 57.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.2 | 7.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 3.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.9 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 4.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 6.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.8 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 35.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.9 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 2.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.0 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 7.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.4 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.1 | 0.5 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.1 | 5.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 11.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 3.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 23.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 2.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 3.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 7.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 0.4 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 3.0 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 3.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 16.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.3 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 3.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.2 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.1 | 2.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 2.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 6.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 14.1 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 4.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 2.0 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 2.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 9.2 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 3.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 1.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 2.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 3.8 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 4.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 1.9 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 1.6 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 6.7 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.7 | GO:0005507 | copper ion binding(GO:0005507) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 16.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 1.1 | 26.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.9 | 50.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.9 | 33.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.8 | 21.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.8 | 129.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.7 | 12.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.6 | 25.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.6 | 10.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.6 | 14.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.5 | 40.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.4 | 32.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.4 | 6.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.4 | 13.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.4 | 8.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.4 | 8.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.4 | 7.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.4 | 21.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.4 | 41.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.3 | 19.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 9.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.3 | 12.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.3 | 10.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 10.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 21.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.3 | 4.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.3 | 13.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.2 | 6.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.2 | 9.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 19.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.2 | 1.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 17.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 4.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 3.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 13.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 7.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 7.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 4.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 5.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 7.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 2.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 4.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 25.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 3.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 7.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 4.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 69.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.6 | 119.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 1.1 | 26.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.1 | 13.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 1.0 | 17.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 1.0 | 11.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.9 | 45.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.9 | 26.2 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.9 | 38.5 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.8 | 31.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.8 | 6.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.8 | 18.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.6 | 5.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.5 | 16.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.5 | 15.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.5 | 7.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.5 | 4.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.5 | 21.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.4 | 62.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.4 | 10.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.4 | 11.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.4 | 9.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 7.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 5.9 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.3 | 5.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.3 | 1.4 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.3 | 5.7 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.3 | 8.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 10.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.3 | 11.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 8.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 3.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.3 | 5.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.3 | 2.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 10.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 3.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 6.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 7.1 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.2 | 4.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 3.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 6.7 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.2 | 7.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 1.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.2 | 4.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 15.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 3.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 2.0 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.2 | 2.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 3.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 3.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 4.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 6.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 4.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 30.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 3.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.4 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 0.8 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 5.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 4.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 3.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 1.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.2 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 4.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 3.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 7.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 3.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 2.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 1.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 2.7 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.2 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.0 | 2.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |