Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
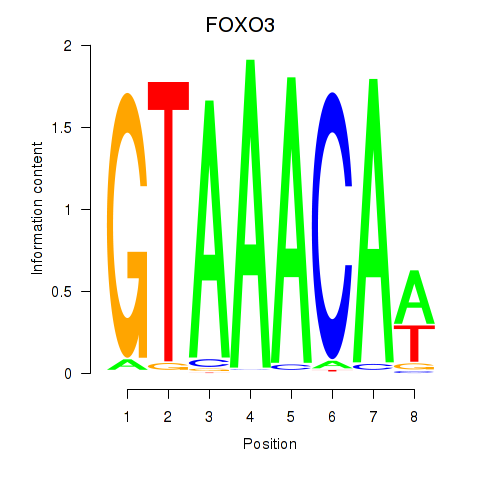
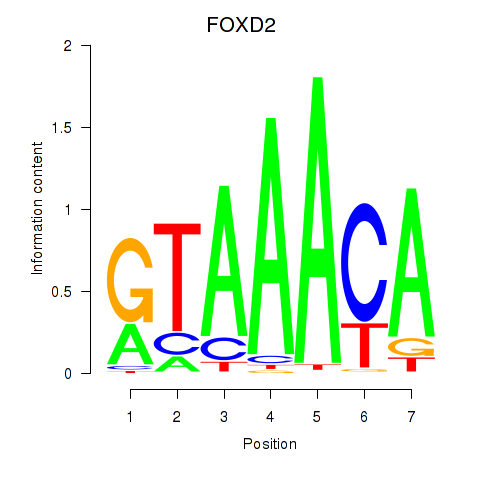
Results for FOXO3_FOXD2
Z-value: 4.76
Transcription factors associated with FOXO3_FOXD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXO3
|
ENSG00000118689.15 | FOXO3 |
|
FOXD2
|
ENSG00000186564.6 | FOXD2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXD2 | hg38_v1_chr1_+_47438036_47438112 | -0.27 | 5.3e-05 | Click! |
| FOXO3 | hg38_v1_chr6_+_108559742_108559835, hg38_v1_chr6_+_108560906_108560970 | -0.15 | 2.8e-02 | Click! |
Activity profile of FOXO3_FOXD2 motif
Sorted Z-values of FOXO3_FOXD2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXO3_FOXD2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.3 | 33.8 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 9.1 | 36.5 | GO:0090095 | metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 6.0 | 30.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 6.0 | 53.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 5.6 | 5.6 | GO:0071335 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 4.7 | 18.8 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 4.0 | 15.8 | GO:0060620 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 3.8 | 15.2 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 3.6 | 10.7 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 3.3 | 50.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 3.2 | 42.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 3.2 | 9.5 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 3.0 | 35.4 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 2.9 | 11.7 | GO:0032600 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 2.7 | 10.8 | GO:0070982 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 2.6 | 15.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 2.5 | 14.7 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 2.4 | 7.1 | GO:0030952 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 2.3 | 21.1 | GO:0070543 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 2.3 | 7.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 2.1 | 6.4 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 2.1 | 8.4 | GO:0043335 | protein unfolding(GO:0043335) |
| 1.9 | 5.8 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 1.9 | 7.7 | GO:2001248 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 1.9 | 11.4 | GO:0044211 | CTP salvage(GO:0044211) |
| 1.8 | 23.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 1.8 | 10.7 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 1.7 | 3.4 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.7 | 6.8 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 1.7 | 10.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 1.7 | 11.8 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 1.7 | 8.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 1.6 | 13.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.6 | 6.5 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 1.6 | 6.5 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 1.6 | 9.7 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 1.6 | 3.2 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 1.6 | 4.8 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 1.6 | 4.7 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 1.6 | 4.7 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 1.5 | 25.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 1.4 | 4.3 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 1.4 | 5.8 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 1.4 | 9.9 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 1.4 | 4.2 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 1.4 | 16.8 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 1.4 | 4.2 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 1.4 | 11.0 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 1.3 | 8.0 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 1.3 | 5.2 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 1.3 | 28.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 1.3 | 3.8 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 1.3 | 5.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 1.2 | 7.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.2 | 28.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 1.2 | 4.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.2 | 5.8 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 1.1 | 9.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 1.1 | 11.2 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 1.1 | 3.3 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 1.1 | 3.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 1.1 | 6.6 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 1.1 | 5.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 1.1 | 5.3 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 1.1 | 3.2 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 1.1 | 9.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 1.1 | 18.9 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 1.1 | 4.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 1.1 | 22.1 | GO:0008228 | opsonization(GO:0008228) |
| 1.0 | 6.2 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 1.0 | 4.0 | GO:0035483 | negative regulation of sodium:proton antiporter activity(GO:0032416) gastric motility(GO:0035482) gastric emptying(GO:0035483) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 1.0 | 8.0 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 1.0 | 13.0 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 1.0 | 7.9 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 1.0 | 6.8 | GO:0030421 | defecation(GO:0030421) |
| 1.0 | 5.8 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 1.0 | 3.8 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.9 | 8.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.9 | 13.1 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.9 | 2.8 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.9 | 3.6 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.9 | 4.5 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.9 | 5.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.9 | 0.9 | GO:1900145 | regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) |
| 0.9 | 2.7 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.9 | 3.5 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.9 | 31.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.9 | 2.6 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.9 | 8.6 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.9 | 15.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.9 | 3.4 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.8 | 3.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.8 | 1.6 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.8 | 0.8 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.8 | 4.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.8 | 9.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.8 | 2.4 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.8 | 18.4 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.8 | 3.0 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.7 | 20.8 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.7 | 3.7 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.7 | 1.4 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.7 | 5.0 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.7 | 4.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.7 | 2.8 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.7 | 2.8 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.7 | 6.9 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.7 | 2.7 | GO:0019376 | galactolipid catabolic process(GO:0019376) |
| 0.7 | 8.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.7 | 12.8 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.7 | 0.7 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.7 | 48.4 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.7 | 4.6 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.6 | 3.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.6 | 3.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.6 | 5.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.6 | 5.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.6 | 17.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.6 | 1.8 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.6 | 1.8 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.6 | 6.5 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.6 | 7.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.6 | 1.2 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.6 | 5.2 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.6 | 2.9 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.6 | 2.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.6 | 1.7 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.6 | 2.8 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.6 | 10.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.5 | 2.7 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.5 | 4.9 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.5 | 3.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.5 | 0.5 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.5 | 2.6 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.5 | 5.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.5 | 3.1 | GO:1904720 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.5 | 2.5 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.5 | 1.5 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.5 | 2.5 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.5 | 5.0 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.5 | 2.5 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.5 | 4.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.5 | 3.9 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.5 | 1.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.5 | 3.9 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.5 | 1.5 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.5 | 1.4 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.5 | 2.9 | GO:1900223 | positive regulation of beta-amyloid clearance(GO:1900223) |
| 0.5 | 6.7 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.5 | 35.0 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.5 | 1.9 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.5 | 30.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.5 | 8.7 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.5 | 3.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.5 | 6.8 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.5 | 1.8 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.4 | 3.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.4 | 3.5 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.4 | 7.0 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.4 | 2.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.4 | 12.2 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.4 | 16.4 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.4 | 2.6 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.4 | 1.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.4 | 2.1 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.4 | 2.6 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.4 | 7.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.4 | 2.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.4 | 4.5 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.4 | 16.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.4 | 2.9 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.4 | 6.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.4 | 1.6 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.4 | 12.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.4 | 1.2 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.4 | 5.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 | 3.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.4 | 1.9 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.4 | 6.2 | GO:0043584 | nose development(GO:0043584) |
| 0.4 | 2.3 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.4 | 1.5 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.4 | 3.7 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.4 | 6.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.4 | 6.9 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.4 | 15.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.4 | 1.4 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.4 | 4.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.4 | 2.1 | GO:0002381 | immunoglobulin production involved in immunoglobulin mediated immune response(GO:0002381) |
| 0.4 | 2.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.4 | 9.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.3 | 3.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 | 4.7 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.3 | 1.7 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.3 | 1.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.3 | 1.9 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.3 | 3.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.3 | 0.6 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.3 | 1.3 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.3 | 3.4 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.3 | 4.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.3 | 1.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.3 | 11.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 | 1.8 | GO:0097460 | ferrous iron import(GO:0070627) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 1.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.3 | 2.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.3 | 4.3 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.3 | 0.9 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 0.3 | 3.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.3 | 6.7 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.3 | 2.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 8.3 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.3 | 2.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.3 | 2.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.3 | 4.5 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.3 | 2.9 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 2.8 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.3 | 11.5 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.3 | 3.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 1.5 | GO:0048840 | otolith development(GO:0048840) |
| 0.3 | 4.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.3 | 4.8 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 1.9 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 6.5 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 | 2.8 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 0.9 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 1.6 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.2 | 7.7 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.2 | 3.7 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.2 | 0.9 | GO:0009956 | radial pattern formation(GO:0009956) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.2 | 4.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 1.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 13.0 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.2 | 1.3 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) UDP-glucuronate metabolic process(GO:0046398) |
| 0.2 | 8.8 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.2 | 4.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 0.8 | GO:0070417 | cellular response to cold(GO:0070417) response to fluoride(GO:1902617) |
| 0.2 | 1.2 | GO:0033632 | negative regulation of extracellular matrix disassembly(GO:0010716) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 | 2.5 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.2 | 15.5 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.2 | 2.5 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.2 | 0.9 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.2 | 11.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 1.6 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.2 | 0.9 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) regulation of serotonin secretion(GO:0014062) |
| 0.2 | 1.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 0.5 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 9.4 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.2 | 2.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 3.5 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.2 | 1.8 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 2.3 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.2 | 1.9 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 1.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 2.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 0.5 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.2 | 2.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 1.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 2.9 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 4.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 2.9 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.2 | 5.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 2.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 3.1 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.1 | 0.3 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 3.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 4.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 1.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.8 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 1.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 11.6 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 1.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.7 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.1 | 0.4 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 2.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.4 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.1 | 0.4 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 1.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 2.0 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 11.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 5.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 2.3 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 1.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 5.1 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.6 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 | 2.5 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 6.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.2 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 2.1 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 2.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.5 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.1 | 14.7 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.1 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.2 | GO:0072537 | fibroblast activation(GO:0072537) |
| 0.1 | 0.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 2.0 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 4.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 4.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 6.0 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.1 | 2.1 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 1.6 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 4.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 8.1 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.1 | 2.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.9 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.1 | 3.3 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 0.2 | GO:0071484 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 0.1 | 7.4 | GO:0060041 | retina development in camera-type eye(GO:0060041) |
| 0.1 | 0.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 1.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 1.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.4 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 1.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 4.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 21.8 | GO:0042493 | response to drug(GO:0042493) |
| 0.1 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 1.5 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.1 | 0.7 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 3.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 3.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 4.1 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.8 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.9 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 1.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 1.1 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.7 | GO:1900274 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.0 | 1.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 1.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 1.0 | GO:0010659 | cardiac muscle cell apoptotic process(GO:0010659) |
| 0.0 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 3.3 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 1.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 2.9 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 1.4 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 1.9 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 2.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 3.7 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 1.0 | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032434) |
| 0.0 | 0.1 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.4 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 3.2 | GO:0022409 | positive regulation of cell-cell adhesion(GO:0022409) |
| 0.0 | 0.7 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.4 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.7 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.2 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.0 | 50.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 9.0 | 53.8 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 3.2 | 42.2 | GO:0042555 | MCM complex(GO:0042555) |
| 2.7 | 10.7 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 2.1 | 8.6 | GO:0044307 | dendritic branch(GO:0044307) |
| 2.0 | 6.1 | GO:0034657 | GID complex(GO:0034657) |
| 2.0 | 6.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 1.9 | 16.8 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 1.6 | 4.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.4 | 4.2 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 1.4 | 6.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 1.2 | 3.6 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 1.0 | 25.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.9 | 14.9 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.9 | 7.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.8 | 3.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.8 | 7.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.8 | 3.9 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.7 | 6.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.7 | 28.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.7 | 33.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.7 | 8.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.7 | 4.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.7 | 6.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.7 | 8.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.7 | 8.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.6 | 10.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.6 | 6.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.6 | 13.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.6 | 4.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.6 | 4.9 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.6 | 2.9 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.6 | 3.4 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.6 | 11.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.6 | 21.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.6 | 12.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.5 | 3.6 | GO:0032021 | NELF complex(GO:0032021) |
| 0.5 | 3.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.5 | 32.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.5 | 4.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.5 | 15.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.5 | 3.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.5 | 2.7 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.4 | 1.8 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.4 | 10.8 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.4 | 8.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.4 | 5.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.4 | 8.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.4 | 2.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.4 | 11.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.4 | 4.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.4 | 2.6 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.4 | 1.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.4 | 2.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 9.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.3 | 5.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 71.7 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.3 | 7.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 6.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.3 | 33.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.3 | 3.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 19.9 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.3 | 4.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 3.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.3 | 1.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 35.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.3 | 6.4 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.3 | 3.2 | GO:0001741 | XY body(GO:0001741) |
| 0.3 | 1.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 2.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 1.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 3.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 2.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 6.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.2 | 4.5 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.2 | 2.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 2.7 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 11.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 0.6 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.2 | 2.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 1.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 1.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 1.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 0.5 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.2 | 15.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 8.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.2 | 11.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 0.7 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 2.6 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 2.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 21.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 3.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 22.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.9 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 6.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 4.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.0 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 2.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.1 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.1 | 0.9 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 2.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 3.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 33.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 11.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 7.6 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 2.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 6.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 3.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 4.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 9.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 4.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 6.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 4.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 4.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 2.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 5.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 8.3 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 3.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 9.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 12.6 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.4 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 3.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 4.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 9.5 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.0 | 4.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 6.1 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 4.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 9.3 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 2.8 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 1.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 3.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 11.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 17.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 30.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 3.2 | 9.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 3.2 | 38.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 3.1 | 33.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 3.0 | 14.9 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 2.8 | 28.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 2.6 | 15.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 2.5 | 14.7 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 2.4 | 18.9 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 2.3 | 7.0 | GO:0031208 | POZ domain binding(GO:0031208) |
| 2.2 | 10.8 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 2.0 | 36.5 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 1.9 | 5.8 | GO:0098519 | polynucleotide 5'-phosphatase activity(GO:0004651) nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 1.9 | 5.8 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 1.9 | 7.7 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 1.9 | 13.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 1.8 | 10.7 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 1.7 | 18.8 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 1.7 | 8.3 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 1.6 | 13.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.6 | 11.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 1.6 | 4.7 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 1.5 | 12.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 1.4 | 5.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 1.4 | 12.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 1.4 | 4.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 1.4 | 4.2 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 1.4 | 6.8 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 1.3 | 9.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 1.3 | 6.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 1.3 | 12.9 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 1.3 | 5.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 1.2 | 7.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 1.2 | 4.8 | GO:0033265 | choline binding(GO:0033265) |
| 1.2 | 35.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 1.2 | 15.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 1.1 | 11.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.1 | 6.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 1.1 | 9.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.0 | 3.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.0 | 4.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.0 | 3.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 1.0 | 11.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 1.0 | 11.8 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 1.0 | 5.8 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 1.0 | 4.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.9 | 25.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.9 | 2.7 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.9 | 6.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.9 | 3.6 | GO:0002046 | opsin binding(GO:0002046) |
| 0.9 | 6.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.9 | 2.6 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.9 | 2.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.9 | 2.6 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.9 | 6.9 | GO:0070697 | activin receptor binding(GO:0070697) type II activin receptor binding(GO:0070699) |
| 0.9 | 2.6 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.9 | 42.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.9 | 3.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.8 | 5.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.8 | 7.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.8 | 7.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.8 | 7.4 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.8 | 3.3 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.8 | 33.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.8 | 3.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.8 | 3.9 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.8 | 8.4 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.8 | 2.3 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.7 | 3.7 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.7 | 5.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.7 | 1.4 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.7 | 9.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.7 | 4.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.7 | 7.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.6 | 6.5 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.6 | 3.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.6 | 5.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.6 | 1.8 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.6 | 3.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.6 | 1.8 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.6 | 1.8 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.6 | 11.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.6 | 4.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.6 | 2.3 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.6 | 1.7 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.6 | 5.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.6 | 3.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.5 | 1.6 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.5 | 2.7 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.5 | 7.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.5 | 7.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.5 | 1.5 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.5 | 3.4 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.5 | 6.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.5 | 1.5 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.5 | 1.9 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.5 | 10.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.5 | 14.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.5 | 2.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.4 | 13.0 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.4 | 3.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.4 | 19.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.4 | 4.3 | GO:0050072 | RNA 7-methylguanosine cap binding(GO:0000340) m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.4 | 16.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.4 | 27.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.4 | 11.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.4 | 8.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 12.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.4 | 5.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.4 | 37.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.4 | 1.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 1.6 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.4 | 47.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.4 | 4.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.4 | 10.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 3.5 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.4 | 6.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 3.7 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.4 | 53.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.4 | 8.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 2.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.4 | 4.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.3 | 9.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.3 | 1.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.3 | 1.6 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 1.9 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.3 | 5.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.3 | 5.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.3 | 6.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 2.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 1.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 1.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 15.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.3 | 5.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.3 | 3.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 4.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.3 | 10.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.3 | 4.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 1.5 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 3.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.3 | 12.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 15.7 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.3 | 1.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 18.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.2 | 0.7 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 6.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 1.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 19.5 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.2 | 8.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 5.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 11.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 4.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 7.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 2.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 3.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 0.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 0.6 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.2 | 3.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 3.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 2.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 6.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 0.6 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 4.6 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.2 | 5.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 0.9 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.2 | 5.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 12.0 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.2 | 4.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 2.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 6.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 28.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 7.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 3.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 14.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.2 | 1.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 1.0 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 2.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 4.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 1.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 1.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 9.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.2 | 4.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.2 | 4.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 4.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 2.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 3.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 8.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 1.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 2.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.0 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 2.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.9 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 2.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.6 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 22.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 5.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 1.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.6 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.1 | 1.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.9 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 3.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 2.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.2 | GO:0004577 | N-acetylglucosaminyldiphosphodolichol N-acetylglucosaminyltransferase activity(GO:0004577) |
| 0.1 | 2.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 4.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.5 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 19.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 12.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 1.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 0.4 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 1.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 14.1 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.1 | 2.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.4 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.1 | 1.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 5.6 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 0.2 | GO:0032557 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) pyrimidine ribonucleotide binding(GO:0032557) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.8 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.5 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 1.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 7.1 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 1.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 3.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 2.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.8 | GO:0005227 | calcium activated cation channel activity(GO:0005227) calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 4.2 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 98.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 1.2 | 30.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 1.0 | 33.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.9 | 66.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.8 | 38.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.7 | 8.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.7 | 15.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.6 | 20.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.6 | 8.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.6 | 30.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.5 | 18.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.5 | 12.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.5 | 19.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.5 | 11.8 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.4 | 16.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.4 | 8.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 20.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.4 | 47.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.4 | 24.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.3 | 41.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.3 | 12.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 4.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.3 | 18.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.3 | 7.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 7.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.3 | 7.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 7.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 11.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.3 | 14.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 11.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 20.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 10.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 7.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 4.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 7.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 6.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 8.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 4.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 3.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 1.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 5.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 17.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 11.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 4.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 10.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 2.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 5.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 1.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 7.6 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 8.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 7.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 2.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 10.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 2.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 5.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 42.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 2.0 | 30.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 1.9 | 11.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 1.4 | 77.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 1.3 | 62.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 1.3 | 18.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 1.2 | 37.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 1.1 | 35.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 1.0 | 28.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 1.0 | 25.3 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.9 | 52.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.9 | 9.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.8 | 20.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.7 | 11.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.7 | 12.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.7 | 9.3 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.7 | 9.8 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.7 | 8.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.7 | 7.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.7 | 14.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.7 | 21.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.6 | 44.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.6 | 10.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.6 | 5.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.6 | 3.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.5 | 15.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.5 | 4.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.5 | 8.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.5 | 13.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.5 | 5.8 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.5 | 11.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.5 | 14.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.4 | 24.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.4 | 4.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.4 | 12.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.4 | 13.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.4 | 18.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.4 | 9.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.4 | 20.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.4 | 7.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.4 | 6.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.4 | 39.5 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.4 | 14.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.4 | 3.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.4 | 8.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.3 | 16.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.3 | 4.7 | REACTOME PURINE METABOLISM | Genes involved in Purine metabolism |
| 0.3 | 6.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.3 | 3.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 5.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.3 | 8.0 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.3 | 1.4 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.3 | 3.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 2.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.3 | 4.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 6.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.3 | 4.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 3.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 6.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 19.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 3.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 4.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 6.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 18.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 2.7 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.2 | 10.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 30.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 1.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.2 | 4.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 1.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.2 | 4.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 2.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 4.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 3.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 4.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 1.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 4.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 5.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 9.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 3.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 7.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 14.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 14.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 2.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.8 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 6.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 13.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 11.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 4.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 2.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 3.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 2.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.8 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 3.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |