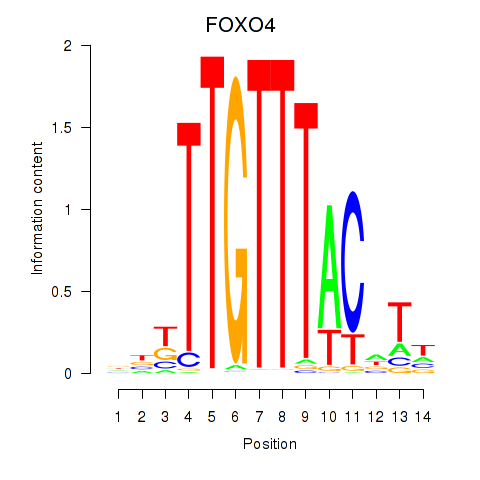
|
chr9_+_127611760
Show fit
|
30.57 |
ENST00000625363.2
ENST00000626539.3
|
STXBP1
|
syntaxin binding protein 1
|
|
chr5_-_42811884
Show fit
|
30.21 |
ENST00000514985.6
ENST00000511224.5
ENST00000507920.5
ENST00000510965.1
|
SELENOP
|
selenoprotein P
|
|
chr5_-_131796921
Show fit
|
29.27 |
ENST00000307968.11
ENST00000307954.12
|
FNIP1
|
folliculin interacting protein 1
|
|
chr5_-_131797030
Show fit
|
26.52 |
ENST00000615660.4
|
FNIP1
|
folliculin interacting protein 1
|
|
chr14_-_59630582
Show fit
|
26.39 |
ENST00000395090.5
|
RTN1
|
reticulon 1
|
|
chr16_+_6019585
Show fit
|
25.58 |
ENST00000547372.5
|
RBFOX1
|
RNA binding fox-1 homolog 1
|
|
chr1_-_182391783
Show fit
|
25.25 |
ENST00000331872.11
ENST00000339526.8
|
GLUL
|
glutamate-ammonia ligase
|
|
chr1_-_182391323
Show fit
|
24.31 |
ENST00000642379.1
|
GLUL
|
glutamate-ammonia ligase
|
|
chr2_-_71227055
Show fit
|
23.65 |
ENST00000244221.9
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chr1_+_92080305
Show fit
|
23.17 |
ENST00000342818.4
ENST00000636805.2
|
BTBD8
|
BTB domain containing 8
|
|
chr11_-_62709493
Show fit
|
23.08 |
ENST00000405837.5
ENST00000531524.5
ENST00000524862.6
ENST00000679883.1
|
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin
|
|
chr1_-_182391363
Show fit
|
22.93 |
ENST00000417584.6
|
GLUL
|
glutamate-ammonia ligase
|
|
chr1_-_56579555
Show fit
|
22.61 |
ENST00000371250.4
|
PLPP3
|
phospholipid phosphatase 3
|
|
chr14_-_103521342
Show fit
|
21.79 |
ENST00000553610.5
|
CKB
|
creatine kinase B
|
|
chr20_+_45406560
Show fit
|
20.53 |
ENST00000372717.5
ENST00000360981.8
|
DBNDD2
|
dysbindin domain containing 2
|
|
chr16_+_6019016
Show fit
|
20.41 |
ENST00000550418.6
|
RBFOX1
|
RNA binding fox-1 homolog 1
|
|
chr19_-_36032799
Show fit
|
19.98 |
ENST00000592017.5
ENST00000360535.9
|
CLIP3
|
CAP-Gly domain containing linker protein 3
|
|
chr16_+_6019071
Show fit
|
18.70 |
ENST00000547605.5
ENST00000553186.5
|
RBFOX1
|
RNA binding fox-1 homolog 1
|
|
chr11_+_114060204
Show fit
|
18.68 |
ENST00000683318.1
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr20_+_45407207
Show fit
|
18.12 |
ENST00000372712.6
|
DBNDD2
|
dysbindin domain containing 2
|
|
chr2_-_2326210
Show fit
|
18.12 |
ENST00000647755.1
|
MYT1L
|
myelin transcription factor 1 like
|
|
chr14_-_21025490
Show fit
|
18.09 |
ENST00000553442.5
ENST00000555869.5
ENST00000556457.5
ENST00000397844.6
ENST00000554415.5
ENST00000556974.5
ENST00000554419.5
ENST00000298687.9
ENST00000397858.5
ENST00000360463.7
ENST00000350792.7
ENST00000397847.6
|
NDRG2
|
NDRG family member 2
|
|
chr19_-_12610799
Show fit
|
17.93 |
ENST00000311437.11
|
ZNF490
|
zinc finger protein 490
|
|
chr17_-_17972374
Show fit
|
16.10 |
ENST00000318094.14
ENST00000540946.5
ENST00000379504.8
ENST00000542206.5
ENST00000395739.8
ENST00000581396.5
ENST00000535933.5
ENST00000579586.1
|
TOM1L2
|
target of myb1 like 2 membrane trafficking protein
|
|
chr2_+_232662733
Show fit
|
15.99 |
ENST00000410095.5
ENST00000611312.1
|
EFHD1
|
EF-hand domain family member D1
|
|
chr16_+_56191476
Show fit
|
15.66 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1
|
|
chr17_-_44324770
Show fit
|
15.39 |
ENST00000592857.5
ENST00000586016.5
ENST00000590194.5
ENST00000588049.5
ENST00000586633.5
ENST00000377095.10
ENST00000537904.6
ENST00000585636.5
ENST00000585523.1
ENST00000225308.12
|
SLC25A39
|
solute carrier family 25 member 39
|
|
chr17_+_17972813
Show fit
|
14.86 |
ENST00000582416.5
ENST00000313838.12
ENST00000399187.6
ENST00000581264.5
ENST00000479684.2
ENST00000584166.5
ENST00000585108.5
ENST00000399182.5
ENST00000579977.1
|
DRC3
|
dynein regulatory complex subunit 3
|
|
chr8_-_133297092
Show fit
|
14.66 |
ENST00000522890.5
ENST00000675983.1
ENST00000518176.5
ENST00000323851.13
ENST00000522476.5
ENST00000518066.5
ENST00000521544.5
ENST00000674605.1
ENST00000518480.5
ENST00000523892.5
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr6_-_152168291
Show fit
|
14.19 |
ENST00000354674.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1
|
|
chr19_-_36528232
Show fit
|
14.04 |
ENST00000592282.1
ENST00000523638.6
|
ZNF260
|
zinc finger protein 260
|
|
chr6_-_152168349
Show fit
|
14.01 |
ENST00000539504.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1
|
|
chr19_+_12610912
Show fit
|
13.98 |
ENST00000446165.2
ENST00000343325.9
|
ZNF791
|
zinc finger protein 791
|
|
chr12_+_79045625
Show fit
|
13.37 |
ENST00000552744.5
|
SYT1
|
synaptotagmin 1
|
|
chr2_-_2326378
Show fit
|
13.16 |
ENST00000647618.1
|
MYT1L
|
myelin transcription factor 1 like
|
|
chr19_+_49877425
Show fit
|
13.13 |
ENST00000622860.4
|
TBC1D17
|
TBC1 domain family member 17
|
|
chr3_-_28348805
Show fit
|
12.64 |
ENST00000457172.5
ENST00000479665.6
|
AZI2
|
5-azacytidine induced 2
|
|
chr1_+_162069768
Show fit
|
12.17 |
ENST00000530878.5
|
NOS1AP
|
nitric oxide synthase 1 adaptor protein
|
|
chr5_-_65624288
Show fit
|
11.93 |
ENST00000381018.7
ENST00000274327.11
ENST00000231524.14
|
TRIM23
|
tripartite motif containing 23
|
|
chr6_+_101398788
Show fit
|
11.85 |
ENST00000369138.5
ENST00000413795.5
ENST00000358361.7
|
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2
|
|
chr19_+_49877660
Show fit
|
11.61 |
ENST00000535102.6
|
TBC1D17
|
TBC1 domain family member 17
|
|
chr18_-_24272179
Show fit
|
11.42 |
ENST00000399443.7
|
OSBPL1A
|
oxysterol binding protein like 1A
|
|
chr2_-_36966471
Show fit
|
11.41 |
ENST00000379213.3
|
STRN
|
striatin
|
|
chr3_+_124384513
Show fit
|
11.03 |
ENST00000682540.1
ENST00000522553.6
ENST00000682695.1
ENST00000682674.1
ENST00000684382.1
|
KALRN
|
kalirin RhoGEF kinase
|
|
chr12_+_12725897
Show fit
|
10.85 |
ENST00000326765.10
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr11_-_114595777
Show fit
|
10.65 |
ENST00000375478.4
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4
|
|
chr11_-_114595750
Show fit
|
10.42 |
ENST00000424261.6
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4
|
|
chr19_+_49877694
Show fit
|
10.05 |
ENST00000221543.10
|
TBC1D17
|
TBC1 domain family member 17
|
|
chr5_-_116536458
Show fit
|
9.74 |
ENST00000510263.5
|
SEMA6A
|
semaphorin 6A
|
|
chr15_-_29822418
Show fit
|
9.73 |
ENST00000614355.5
ENST00000495972.6
ENST00000346128.10
|
TJP1
|
tight junction protein 1
|
|
chr15_+_43692886
Show fit
|
9.63 |
ENST00000434505.5
ENST00000411750.5
|
CKMT1A
|
creatine kinase, mitochondrial 1A
|
|
chr1_+_43389889
Show fit
|
9.61 |
ENST00000562955.2
ENST00000634258.3
|
SZT2
|
SZT2 subunit of KICSTOR complex
|
|
chr4_-_86358487
Show fit
|
9.54 |
ENST00000641066.1
ENST00000512689.6
ENST00000641803.1
ENST00000640970.1
ENST00000641983.1
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr1_+_150257247
Show fit
|
9.30 |
ENST00000647854.1
|
CA14
|
carbonic anhydrase 14
|
|
chr18_-_55635948
Show fit
|
9.24 |
ENST00000565124.4
ENST00000398339.5
|
TCF4
|
transcription factor 4
|
|
chr5_-_108367860
Show fit
|
9.19 |
ENST00000496714.2
|
FBXL17
|
F-box and leucine rich repeat protein 17
|
|
chr1_+_162069674
Show fit
|
9.09 |
ENST00000361897.10
|
NOS1AP
|
nitric oxide synthase 1 adaptor protein
|
|
chr11_-_56292254
Show fit
|
8.95 |
ENST00000641600.1
|
OR8H1
|
olfactory receptor family 8 subfamily H member 1
|
|
chr17_+_79778135
Show fit
|
8.92 |
ENST00000310942.9
ENST00000269399.5
|
CBX2
|
chromobox 2
|
|
chr21_-_31558977
Show fit
|
8.85 |
ENST00000286827.7
ENST00000541036.5
|
TIAM1
|
TIAM Rac1 associated GEF 1
|
|
chr7_+_20615653
Show fit
|
8.76 |
ENST00000404938.7
|
ABCB5
|
ATP binding cassette subfamily B member 5
|
|
chr10_+_110225955
Show fit
|
8.72 |
ENST00000239007.11
|
MXI1
|
MAX interactor 1, dimerization protein
|
|
chr21_-_31160904
Show fit
|
8.70 |
ENST00000636887.1
|
TIAM1
|
TIAM Rac1 associated GEF 1
|
|
chr15_+_43593054
Show fit
|
8.60 |
ENST00000453782.5
ENST00000300283.10
ENST00000437924.5
|
CKMT1B
|
creatine kinase, mitochondrial 1B
|
|
chr14_+_94110728
Show fit
|
8.56 |
ENST00000616764.4
ENST00000618863.1
ENST00000611954.4
ENST00000618200.4
ENST00000621160.4
ENST00000555819.5
ENST00000620396.4
ENST00000612813.4
ENST00000620066.1
|
IFI27
|
interferon alpha inducible protein 27
|
|
chr19_-_40226682
Show fit
|
8.41 |
ENST00000430325.7
ENST00000599263.6
|
CCNP
|
cyclin P
|
|
chr2_-_112255015
Show fit
|
8.31 |
ENST00000615791.1
ENST00000409573.7
ENST00000272570.9
|
ZC3H8
|
zinc finger CCCH-type containing 8
|
|
chr12_-_12338674
Show fit
|
8.22 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1
|
|
chr7_+_134866831
Show fit
|
8.21 |
ENST00000435928.1
|
CALD1
|
caldesmon 1
|
|
chr14_+_70452161
Show fit
|
8.11 |
ENST00000603540.2
|
ADAM21
|
ADAM metallopeptidase domain 21
|
|
chr6_-_46921926
Show fit
|
8.08 |
ENST00000283296.12
|
ADGRF5
|
adhesion G protein-coupled receptor F5
|
|
chr4_-_185775271
Show fit
|
8.04 |
ENST00000430503.5
ENST00000319454.10
ENST00000450341.5
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr11_+_46617521
Show fit
|
8.02 |
ENST00000580238.5
ENST00000581416.5
ENST00000529655.5
ENST00000533325.5
ENST00000683050.1
ENST00000581438.5
ENST00000583249.5
ENST00000530500.5
ENST00000526508.5
ENST00000578626.5
ENST00000577256.5
ENST00000524625.5
ENST00000582547.5
ENST00000359513.8
ENST00000528494.5
|
ATG13
|
autophagy related 13
|
|
chr4_-_89836213
Show fit
|
7.91 |
ENST00000618500.4
ENST00000508895.5
|
SNCA
|
synuclein alpha
|
|
chr2_+_235494024
Show fit
|
7.90 |
ENST00000304032.13
ENST00000409457.5
ENST00000336665.9
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr10_+_91220603
Show fit
|
7.86 |
ENST00000336126.6
|
PCGF5
|
polycomb group ring finger 5
|
|
chr3_+_159852933
Show fit
|
7.57 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr3_-_139539577
Show fit
|
7.52 |
ENST00000619087.4
|
RBP1
|
retinol binding protein 1
|
|
chr2_-_165204042
Show fit
|
7.50 |
ENST00000283254.12
ENST00000453007.1
|
SCN3A
|
sodium voltage-gated channel alpha subunit 3
|
|
chr8_-_42843201
Show fit
|
7.40 |
ENST00000529779.1
ENST00000345117.2
ENST00000254250.7
|
THAP1
|
THAP domain containing 1
|
|
chr17_+_3475959
Show fit
|
7.38 |
ENST00000263080.3
|
ASPA
|
aspartoacylase
|
|
chr2_-_2331225
Show fit
|
7.22 |
ENST00000648627.1
ENST00000649663.1
ENST00000650560.1
ENST00000428368.7
ENST00000648316.1
ENST00000648665.1
ENST00000649313.1
ENST00000399161.7
ENST00000647738.2
|
MYT1L
|
myelin transcription factor 1 like
|
|
chr6_-_167157980
Show fit
|
7.14 |
ENST00000366834.2
|
GPR31
|
G protein-coupled receptor 31
|
|
chr5_-_218136
Show fit
|
7.10 |
ENST00000296824.4
|
CCDC127
|
coiled-coil domain containing 127
|
|
chr3_+_138348823
Show fit
|
7.07 |
ENST00000475711.5
ENST00000464896.5
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr2_+_32277883
Show fit
|
6.93 |
ENST00000238831.9
|
YIPF4
|
Yip1 domain family member 4
|
|
chr3_+_138349209
Show fit
|
6.92 |
ENST00000474559.1
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr10_-_60141004
Show fit
|
6.79 |
ENST00000355288.6
|
ANK3
|
ankyrin 3
|
|
chr6_+_32154131
Show fit
|
6.78 |
ENST00000375143.6
ENST00000324816.11
ENST00000424499.1
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr4_+_123399488
Show fit
|
6.74 |
ENST00000394339.2
|
SPRY1
|
sprouty RTK signaling antagonist 1
|
|
chr5_-_134371004
Show fit
|
6.74 |
ENST00000521755.1
ENST00000523054.5
ENST00000518409.1
|
CDKL3
ENSG00000273345.5
|
cyclin dependent kinase like 3
novel transcript
|
|
chr2_-_165203870
Show fit
|
6.71 |
ENST00000639244.1
ENST00000409101.7
ENST00000668657.1
|
SCN3A
|
sodium voltage-gated channel alpha subunit 3
|
|
chr12_-_10098977
Show fit
|
6.70 |
ENST00000315330.8
ENST00000457018.6
|
CLEC1A
|
C-type lectin domain family 1 member A
|
|
chr19_+_41262656
Show fit
|
6.68 |
ENST00000599719.5
ENST00000601309.5
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U like 1
|
|
chr9_+_27109200
Show fit
|
6.59 |
ENST00000380036.10
|
TEK
|
TEK receptor tyrosine kinase
|
|
chr10_-_50885619
Show fit
|
6.49 |
ENST00000373997.8
|
A1CF
|
APOBEC1 complementation factor
|
|
chr14_-_75051426
Show fit
|
6.46 |
ENST00000556257.5
ENST00000355774.7
ENST00000557648.1
ENST00000553263.1
ENST00000380968.6
|
MLH3
|
mutL homolog 3
|
|
chr4_+_87832917
Show fit
|
6.32 |
ENST00000395102.8
ENST00000497649.6
ENST00000540395.1
ENST00000560249.5
ENST00000511670.5
ENST00000361056.3
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chr4_-_185775890
Show fit
|
6.18 |
ENST00000437304.6
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr11_-_66907891
Show fit
|
6.14 |
ENST00000393955.6
|
PC
|
pyruvate carboxylase
|
|
chr17_-_8118489
Show fit
|
6.13 |
ENST00000380149.6
ENST00000448843.7
|
ALOXE3
|
arachidonate lipoxygenase 3
|
|
chr19_-_58150772
Show fit
|
6.09 |
ENST00000597186.5
ENST00000598312.6
|
ZNF329
|
zinc finger protein 329
|
|
chr8_-_71547626
Show fit
|
6.07 |
ENST00000647540.1
ENST00000644229.1
|
EYA1
|
EYA transcriptional coactivator and phosphatase 1
|
|
chr9_+_27109135
Show fit
|
6.06 |
ENST00000519097.5
ENST00000615002.4
|
TEK
|
TEK receptor tyrosine kinase
|
|
chr7_-_712437
Show fit
|
6.04 |
ENST00000360274.8
|
PRKAR1B
|
protein kinase cAMP-dependent type I regulatory subunit beta
|
|
chr6_+_122789197
Show fit
|
6.01 |
ENST00000368440.5
|
SMPDL3A
|
sphingomyelin phosphodiesterase acid like 3A
|
|
chr19_+_15049469
Show fit
|
5.99 |
ENST00000427043.4
|
CASP14
|
caspase 14
|
|
chr11_+_57598184
Show fit
|
5.98 |
ENST00000677625.1
ENST00000676670.1
|
SERPING1
|
serpin family G member 1
|
|
chr1_-_2003780
Show fit
|
5.89 |
ENST00000682832.1
|
CFAP74
|
cilia and flagella associated protein 74
|
|
chr18_+_34493428
Show fit
|
5.86 |
ENST00000682483.1
|
DTNA
|
dystrobrevin alpha
|
|
chr9_+_27109393
Show fit
|
5.83 |
ENST00000406359.8
|
TEK
|
TEK receptor tyrosine kinase
|
|
chr11_+_57597563
Show fit
|
5.81 |
ENST00000619430.2
ENST00000457869.1
ENST00000340687.10
ENST00000278407.9
ENST00000378323.8
ENST00000378324.6
ENST00000403558.1
|
SERPING1
|
serpin family G member 1
|
|
chr1_+_151766655
Show fit
|
5.78 |
ENST00000400999.7
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr3_+_148730100
Show fit
|
5.77 |
ENST00000474935.5
ENST00000475347.5
ENST00000461609.1
|
AGTR1
|
angiotensin II receptor type 1
|
|
chr10_-_50885656
Show fit
|
5.70 |
ENST00000374001.6
ENST00000395489.6
ENST00000282641.6
ENST00000395495.5
ENST00000373995.7
ENST00000414883.1
|
A1CF
|
APOBEC1 complementation factor
|
|
chr16_-_4273014
Show fit
|
5.68 |
ENST00000204517.11
|
TFAP4
|
transcription factor AP-4
|
|
chr2_+_174395721
Show fit
|
5.63 |
ENST00000272732.11
ENST00000458563.5
ENST00000409673.7
ENST00000435964.1
ENST00000424069.5
ENST00000427038.5
|
SCRN3
|
secernin 3
|
|
chr2_+_65056382
Show fit
|
5.54 |
ENST00000377990.7
ENST00000537589.1
ENST00000260569.4
|
CEP68
|
centrosomal protein 68
|
|
chr15_-_55365231
Show fit
|
5.54 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1
|
|
chr2_+_227616998
Show fit
|
5.44 |
ENST00000641801.1
|
SCYGR4
|
small cysteine and glycine repeat containing 4
|
|
chr10_-_60572599
Show fit
|
5.34 |
ENST00000503366.5
|
ANK3
|
ankyrin 3
|
|
chr6_+_32154010
Show fit
|
5.32 |
ENST00000375137.6
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr19_+_1205761
Show fit
|
5.30 |
ENST00000326873.12
ENST00000586243.5
|
STK11
|
serine/threonine kinase 11
|
|
chr11_+_112961247
Show fit
|
5.25 |
ENST00000621518.4
ENST00000618266.4
ENST00000615112.4
ENST00000615285.4
ENST00000619839.4
ENST00000401611.6
ENST00000621128.4
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr9_-_15472732
Show fit
|
5.20 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr5_-_10307821
Show fit
|
5.20 |
ENST00000296658.4
|
CMBL
|
carboxymethylenebutenolidase homolog
|
|
chr16_+_1680337
Show fit
|
5.17 |
ENST00000566691.5
ENST00000382710.8
|
JPT2
|
Jupiter microtubule associated homolog 2
|
|
chr10_+_80413817
Show fit
|
5.11 |
ENST00000372187.9
|
PRXL2A
|
peroxiredoxin like 2A
|
|
chr3_-_186109067
Show fit
|
5.09 |
ENST00000306376.10
|
ETV5
|
ETS variant transcription factor 5
|
|
chr10_+_122163590
Show fit
|
5.08 |
ENST00000368999.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2
|
|
chr1_+_110210272
Show fit
|
5.04 |
ENST00000438661.3
|
KCNC4
|
potassium voltage-gated channel subfamily C member 4
|
|
chr13_-_98977975
Show fit
|
4.99 |
ENST00000376460.5
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr7_+_106865263
Show fit
|
4.87 |
ENST00000440650.6
ENST00000496166.6
ENST00000473541.5
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma
|
|
chr15_-_58279245
Show fit
|
4.87 |
ENST00000558231.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2
|
|
chr18_+_34493386
Show fit
|
4.85 |
ENST00000679936.1
|
DTNA
|
dystrobrevin alpha
|
|
chr4_+_95051671
Show fit
|
4.70 |
ENST00000440890.7
|
BMPR1B
|
bone morphogenetic protein receptor type 1B
|
|
chr2_-_182866627
Show fit
|
4.69 |
ENST00000295113.5
|
FRZB
|
frizzled related protein
|
|
chr11_-_94493798
Show fit
|
4.67 |
ENST00000538923.1
ENST00000540013.5
ENST00000407439.7
ENST00000323929.8
ENST00000393241.8
|
MRE11
|
MRE11 homolog, double strand break repair nuclease
|
|
chr3_-_45884685
Show fit
|
4.63 |
ENST00000684620.1
|
LZTFL1
|
leucine zipper transcription factor like 1
|
|
chr13_-_98978022
Show fit
|
4.59 |
ENST00000682017.1
ENST00000442173.5
ENST00000627024.2
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr11_+_77821105
Show fit
|
4.43 |
ENST00000532481.5
|
AAMDC
|
adipogenesis associated Mth938 domain containing
|
|
chr4_-_89835617
Show fit
|
4.40 |
ENST00000611107.1
ENST00000345009.8
ENST00000505199.5
ENST00000502987.5
|
SNCA
|
synuclein alpha
|
|
chr8_-_42768781
Show fit
|
4.33 |
ENST00000276410.7
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit
|
|
chr5_-_131796965
Show fit
|
4.32 |
ENST00000514667.1
ENST00000511848.1
ENST00000510461.6
|
ENSG00000273217.1
FNIP1
|
novel protein
folliculin interacting protein 1
|
|
chr5_+_80407994
Show fit
|
4.28 |
ENST00000338008.9
ENST00000510158.5
|
ZFYVE16
|
zinc finger FYVE-type containing 16
|
|
chr17_-_42745025
Show fit
|
4.24 |
ENST00000592492.5
ENST00000585893.5
ENST00000593214.5
ENST00000590078.5
ENST00000428826.7
ENST00000586382.5
ENST00000415827.6
ENST00000592743.5
ENST00000586089.5
|
EZH1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit
|
|
chr18_+_62715526
Show fit
|
4.23 |
ENST00000262719.10
|
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1
|
|
chr1_-_153541765
Show fit
|
4.22 |
ENST00000368718.5
|
S100A5
|
S100 calcium binding protein A5
|
|
chr7_-_13988863
Show fit
|
4.22 |
ENST00000405358.8
|
ETV1
|
ETS variant transcription factor 1
|
|
chr1_-_56966006
Show fit
|
4.19 |
ENST00000371237.9
|
C8B
|
complement C8 beta chain
|
|
chr5_+_80408031
Show fit
|
4.15 |
ENST00000505560.5
ENST00000509562.2
|
ZFYVE16
|
zinc finger FYVE-type containing 16
|
|
chr4_-_152411734
Show fit
|
3.99 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr2_+_156436345
Show fit
|
3.96 |
ENST00000438166.7
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2
|
|
chr7_-_95596507
Show fit
|
3.88 |
ENST00000005178.6
|
PDK4
|
pyruvate dehydrogenase kinase 4
|
|
chr4_-_69961007
Show fit
|
3.84 |
ENST00000353151.3
|
CSN2
|
casein beta
|
|
chr14_-_99272184
Show fit
|
3.78 |
ENST00000357195.8
|
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B
|
|
chr14_-_70809494
Show fit
|
3.77 |
ENST00000381250.8
ENST00000554752.7
ENST00000555993.6
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr3_-_46566276
Show fit
|
3.77 |
ENST00000395905.8
|
LRRC2
|
leucine rich repeat containing 2
|
|
chr20_-_46364385
Show fit
|
3.71 |
ENST00000243896.6
ENST00000543605.5
ENST00000372230.10
ENST00000317734.12
|
SLC35C2
|
solute carrier family 35 member C2
|
|
chr2_+_172556039
Show fit
|
3.70 |
ENST00000410055.5
ENST00000282077.8
|
PDK1
|
pyruvate dehydrogenase kinase 1
|
|
chr13_+_97434154
Show fit
|
3.68 |
ENST00000245304.5
|
RAP2A
|
RAP2A, member of RAS oncogene family
|
|
chr3_+_108296489
Show fit
|
3.68 |
ENST00000619531.4
|
HHLA2
|
HERV-H LTR-associating 2
|
|
chr1_+_96722628
Show fit
|
3.61 |
ENST00000675401.1
|
PTBP2
|
polypyrimidine tract binding protein 2
|
|
chr10_+_97640686
Show fit
|
3.60 |
ENST00000370631.3
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chr17_+_74431338
Show fit
|
3.60 |
ENST00000342648.9
ENST00000652232.1
ENST00000481232.2
|
GPRC5C
|
G protein-coupled receptor class C group 5 member C
|
|
chr5_-_126595185
Show fit
|
3.59 |
ENST00000510111.6
ENST00000635851.1
ENST00000637964.1
ENST00000413020.6
ENST00000637782.1
ENST00000409134.8
ENST00000637272.1
ENST00000636879.1
ENST00000636743.1
ENST00000636886.1
ENST00000509270.2
|
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1
|
|
chr1_-_168137430
Show fit
|
3.50 |
ENST00000546300.5
ENST00000271357.9
ENST00000367835.1
ENST00000537209.5
|
GPR161
|
G protein-coupled receptor 161
|
|
chr2_-_36966503
Show fit
|
3.48 |
ENST00000263918.9
|
STRN
|
striatin
|
|
chr8_-_42768602
Show fit
|
3.45 |
ENST00000534622.5
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit
|
|
chr7_-_38349972
Show fit
|
3.39 |
ENST00000390344.2
|
TRGV5
|
T cell receptor gamma variable 5
|
|
chr10_+_122163672
Show fit
|
3.33 |
ENST00000369004.7
ENST00000260733.7
|
TACC2
|
transforming acidic coiled-coil containing protein 2
|
|
chr12_-_70637405
Show fit
|
3.29 |
ENST00000548122.2
ENST00000551525.5
ENST00000550358.5
ENST00000334414.11
|
PTPRB
|
protein tyrosine phosphatase receptor type B
|
|
chr11_-_5269933
Show fit
|
3.29 |
ENST00000396895.3
|
HBE1
|
hemoglobin subunit epsilon 1
|
|
chr7_+_135148041
Show fit
|
3.21 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140
|
|
chr7_+_106865474
Show fit
|
3.18 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma
|
|
chr8_-_40897814
Show fit
|
3.11 |
ENST00000297737.11
ENST00000315769.11
|
ZMAT4
|
zinc finger matrin-type 4
|
|
chr17_-_69141878
Show fit
|
3.09 |
ENST00000590645.1
ENST00000284425.7
|
ABCA6
|
ATP binding cassette subfamily A member 6
|
|
chr18_-_55402187
Show fit
|
3.09 |
ENST00000630268.2
ENST00000570177.6
|
TCF4
|
transcription factor 4
|
|
chr7_+_99828010
Show fit
|
2.96 |
ENST00000631161.2
ENST00000354829.7
ENST00000342499.8
ENST00000417625.5
ENST00000415413.5
ENST00000444905.5
ENST00000222382.5
ENST00000312017.9
|
CYP3A43
|
cytochrome P450 family 3 subfamily A member 43
|
|
chr10_+_122163426
Show fit
|
2.88 |
ENST00000360561.7
|
TACC2
|
transforming acidic coiled-coil containing protein 2
|
|
chr9_+_706841
Show fit
|
2.88 |
ENST00000382293.7
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr8_-_53843228
Show fit
|
2.83 |
ENST00000359530.7
|
ATP6V1H
|
ATPase H+ transporting V1 subunit H
|
|
chr6_-_117425855
Show fit
|
2.81 |
ENST00000368508.7
|
ROS1
|
ROS proto-oncogene 1, receptor tyrosine kinase
|
|
chr15_-_30991415
Show fit
|
2.76 |
ENST00000563714.5
|
MTMR10
|
myotubularin related protein 10
|
|
chrX_-_149595314
Show fit
|
2.74 |
ENST00000598963.3
|
HSFX2
|
heat shock transcription factor family, X-linked 2
|
|
chr16_+_28846674
Show fit
|
2.74 |
ENST00000322610.12
|
SH2B1
|
SH2B adaptor protein 1
|
|
chr4_-_108166715
Show fit
|
2.69 |
ENST00000510624.5
|
LEF1
|
lymphoid enhancer binding factor 1
|
|
chr9_+_5510492
Show fit
|
2.67 |
ENST00000397747.5
|
PDCD1LG2
|
programmed cell death 1 ligand 2
|
|
chr1_-_227318125
Show fit
|
2.66 |
ENST00000366764.7
|
CDC42BPA
|
CDC42 binding protein kinase alpha
|
|
chr8_+_119067239
Show fit
|
2.64 |
ENST00000332843.3
|
COLEC10
|
collectin subfamily member 10
|
|
chr1_+_160796157
Show fit
|
2.64 |
ENST00000263285.11
ENST00000368039.2
|
LY9
|
lymphocyte antigen 9
|
|
chr10_+_110207587
Show fit
|
2.62 |
ENST00000332674.9
ENST00000453116.5
|
MXI1
|
MAX interactor 1, dimerization protein
|
|
chr10_-_1200360
Show fit
|
2.59 |
ENST00000381310.7
ENST00000381305.1
|
ADARB2
|
adenosine deaminase RNA specific B2 (inactive)
|
|
chr11_-_123741614
Show fit
|
2.57 |
ENST00000526252.5
ENST00000530393.6
ENST00000533463.5
ENST00000336139.8
ENST00000529691.1
ENST00000528306.5
|
ZNF202
|
zinc finger protein 202
|
|
chr1_-_56966133
Show fit
|
2.54 |
ENST00000535057.5
ENST00000543257.5
|
C8B
|
complement C8 beta chain
|
|
chr8_-_86743626
Show fit
|
2.54 |
ENST00000320005.6
|
CNGB3
|
cyclic nucleotide gated channel subunit beta 3
|
|
chr20_-_44521989
Show fit
|
2.53 |
ENST00000342374.5
ENST00000255175.5
|
SERINC3
|
serine incorporator 3
|
|
chr12_-_102478539
Show fit
|
2.53 |
ENST00000424202.6
|
IGF1
|
insulin like growth factor 1
|
|
chr14_-_92106535
Show fit
|
2.53 |
ENST00000526872.3
ENST00000532032.5
ENST00000506466.5
ENST00000554592.5
ENST00000555381.5
ENST00000553491.5
ENST00000556220.5
ENST00000557311.6
ENST00000617719.4
ENST00000554672.6
ENST00000644486.2
ENST00000502250.5
ENST00000503767.5
|
ATXN3
|
ataxin 3
|
|
chr16_+_77199408
Show fit
|
2.51 |
ENST00000378644.5
|
SYCE1L
|
synaptonemal complex central element protein 1 like
|