Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
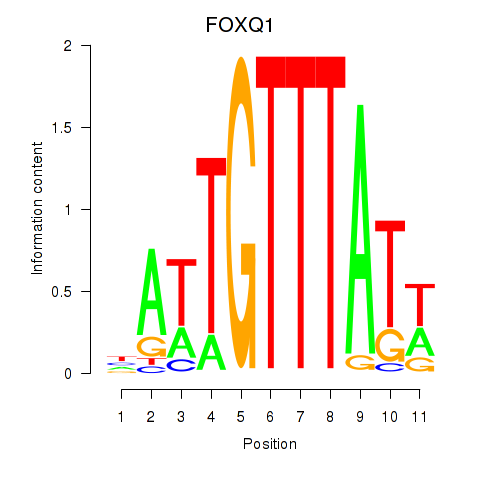
Results for FOXQ1
Z-value: 6.59
Transcription factors associated with FOXQ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXQ1
|
ENSG00000164379.7 | FOXQ1 |
Activity profile of FOXQ1 motif
Sorted Z-values of FOXQ1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXQ1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.8 | 29.5 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 7.4 | 29.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 4.5 | 22.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 3.7 | 11.0 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 3.4 | 23.5 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 3.3 | 10.0 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 3.2 | 25.6 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 3.1 | 12.5 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 3.0 | 15.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 2.7 | 8.2 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 2.7 | 8.2 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 2.6 | 15.4 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 2.5 | 12.6 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 2.4 | 7.3 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 2.3 | 6.9 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 2.2 | 20.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 2.2 | 6.6 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 2.1 | 10.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 2.0 | 11.9 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 1.9 | 5.8 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 1.9 | 13.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.8 | 11.0 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 1.8 | 27.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.7 | 15.7 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 1.7 | 5.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 1.7 | 5.0 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 1.6 | 8.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 1.6 | 7.9 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 1.6 | 6.3 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 1.6 | 4.7 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 1.4 | 5.7 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 1.4 | 4.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 1.3 | 5.4 | GO:0051919 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 1.3 | 18.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 1.2 | 10.7 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 1.1 | 6.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 1.1 | 11.7 | GO:0051451 | myoblast migration(GO:0051451) |
| 1.0 | 8.8 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 1.0 | 6.8 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 1.0 | 16.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 1.0 | 19.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.9 | 3.6 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.9 | 7.0 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.9 | 3.5 | GO:0060437 | lung growth(GO:0060437) |
| 0.9 | 4.3 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.8 | 3.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.8 | 3.9 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.8 | 6.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.7 | 3.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.7 | 3.6 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.7 | 20.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.7 | 10.8 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.7 | 2.8 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.7 | 3.4 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.7 | 4.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.7 | 4.7 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.7 | 7.9 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.7 | 8.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.6 | 8.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.6 | 3.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.6 | 8.2 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.6 | 4.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.6 | 2.5 | GO:0061643 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.6 | 6.5 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.6 | 9.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.6 | 4.5 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.6 | 5.7 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.5 | 2.2 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.5 | 1.6 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.5 | 11.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.5 | 8.3 | GO:1903764 | regulation of potassium ion import(GO:1903286) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.5 | 8.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.5 | 2.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.5 | 6.1 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.5 | 3.0 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.5 | 9.8 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.4 | 1.8 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.4 | 14.9 | GO:0046655 | tetrahydrofolate metabolic process(GO:0046653) folic acid metabolic process(GO:0046655) |
| 0.4 | 3.7 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 1.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.4 | 2.8 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.4 | 7.9 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.4 | 3.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.4 | 11.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.4 | 2.3 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.4 | 3.8 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.4 | 2.6 | GO:1990822 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.4 | 3.4 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.4 | 21.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.4 | 4.9 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.4 | 8.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.4 | 4.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 1.7 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.3 | 13.7 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.3 | 3.5 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.3 | 10.9 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.3 | 1.2 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.3 | 4.9 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.3 | 10.7 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 1.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.3 | 3.9 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.3 | 8.0 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.3 | 4.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.3 | 1.4 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.3 | 8.1 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.2 | 6.0 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.2 | 0.9 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 4.6 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.2 | 2.7 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.2 | 1.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.2 | 14.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 4.6 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.2 | 1.1 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.2 | 1.6 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 5.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.2 | 16.7 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.2 | 12.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 3.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 3.9 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 6.3 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.2 | 1.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 1.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 0.7 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 | 1.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 1.6 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.9 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 8.7 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 6.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.0 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 3.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 10.2 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.1 | 1.0 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 10.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 4.3 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of adherens junction organization(GO:1903393) |
| 0.1 | 5.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 4.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 3.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 4.3 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 4.9 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 6.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 2.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 1.1 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.1 | 1.1 | GO:2000096 | segment specification(GO:0007379) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 3.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 3.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 10.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 2.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.8 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 10.7 | GO:0006302 | double-strand break repair(GO:0006302) |
| 0.1 | 0.9 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 5.6 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 3.1 | GO:0072413 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 | 1.9 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 5.3 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 6.0 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 9.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 1.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.2 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 3.2 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.4 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 2.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 2.8 | GO:0050679 | positive regulation of epithelial cell proliferation(GO:0050679) |
| 0.0 | 1.3 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 1.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.5 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 3.7 | 29.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 3.4 | 23.5 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 3.3 | 10.0 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 2.6 | 7.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.2 | 26.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 1.7 | 7.0 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 1.7 | 16.7 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 1.2 | 13.7 | GO:0043203 | axon hillock(GO:0043203) |
| 1.2 | 6.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 1.0 | 8.2 | GO:0070187 | telosome(GO:0070187) |
| 1.0 | 9.8 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.9 | 15.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.8 | 17.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.8 | 6.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.7 | 5.9 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.6 | 6.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.6 | 3.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.6 | 4.3 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.6 | 44.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.5 | 3.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.5 | 2.7 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.5 | 7.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.5 | 10.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.5 | 8.3 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.5 | 7.8 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.5 | 1.4 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.5 | 2.7 | GO:0002177 | manchette(GO:0002177) |
| 0.4 | 4.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.4 | 8.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.3 | 6.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 1.6 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.3 | 8.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.3 | 6.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 2.7 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 13.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 25.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 1.0 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.2 | 1.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 6.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 14.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 12.5 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 2.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.2 | 4.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 2.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 4.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 1.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.2 | 9.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 1.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 1.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 3.4 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.2 | 10.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 6.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 13.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 4.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 16.0 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 24.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 25.5 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 2.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 8.5 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 12.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 9.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 2.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.5 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 3.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 10.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 16.0 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 11.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 1.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 9.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 10.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 40.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 9.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 6.3 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 17.4 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 4.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 14.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 6.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 10.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 4.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 13.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.7 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 17.4 | GO:0044429 | mitochondrial part(GO:0044429) |
| 0.0 | 13.5 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.8 | 29.5 | GO:0031208 | POZ domain binding(GO:0031208) |
| 5.7 | 22.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 3.8 | 15.2 | GO:0002060 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 3.7 | 14.9 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 3.6 | 10.7 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 3.2 | 9.7 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 3.2 | 25.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 3.0 | 9.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 2.8 | 16.7 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 2.7 | 13.7 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 2.7 | 8.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 2.3 | 6.9 | GO:0047787 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 2.2 | 6.6 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 2.0 | 6.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 1.9 | 7.6 | GO:0033265 | choline binding(GO:0033265) |
| 1.9 | 5.7 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 1.8 | 11.0 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 1.7 | 11.9 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.7 | 5.0 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 1.6 | 4.9 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 1.4 | 17.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 1.4 | 15.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 1.4 | 8.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.3 | 3.9 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 1.3 | 3.9 | GO:0003860 | 3-hydroxyisobutyryl-CoA hydrolase activity(GO:0003860) |
| 1.3 | 3.8 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 1.3 | 10.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.1 | 9.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 1.0 | 3.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 1.0 | 3.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 1.0 | 4.9 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 1.0 | 20.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.9 | 3.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.9 | 6.8 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.8 | 4.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.8 | 7.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.8 | 6.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.8 | 3.0 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.7 | 5.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.7 | 5.8 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.7 | 4.3 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.7 | 5.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.7 | 27.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.6 | 14.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.6 | 5.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.6 | 27.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.6 | 11.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.5 | 10.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.5 | 4.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.5 | 5.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.5 | 16.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.5 | 3.9 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.5 | 15.8 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.5 | 8.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.5 | 3.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.5 | 16.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.4 | 7.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 1.6 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.4 | 1.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.4 | 1.6 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.4 | 2.8 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.4 | 8.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.4 | 11.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.4 | 3.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.4 | 5.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 12.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.4 | 2.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.4 | 24.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.4 | 2.5 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.3 | 6.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.3 | 4.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.3 | 19.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.3 | 1.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 20.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.3 | 14.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 5.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 6.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 1.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 6.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 2.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 8.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 5.8 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 15.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 2.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 2.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 1.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 1.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 2.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 6.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 33.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 2.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 1.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.2 | 1.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 3.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 5.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 3.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 4.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 9.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 18.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 1.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.1 | 1.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 2.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 7.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 4.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.4 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 12.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 30.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 1.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 1.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 5.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 12.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 3.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 5.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 2.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 6.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 29.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.4 | 19.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.4 | 8.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.4 | 14.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.4 | 12.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 14.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.3 | 3.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.3 | 18.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.3 | 13.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 23.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 13.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 8.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 6.1 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.2 | 7.5 | ST ADRENERGIC | Adrenergic Pathway |
| 0.2 | 8.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 11.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 11.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 13.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 9.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 7.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 4.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 8.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 2.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 3.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 7.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 7.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 8.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 6.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.3 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 8.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 8.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 7.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 2.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 4.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 4.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 11.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 6.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 1.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 2.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 2.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 3.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 26.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 1.5 | 22.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 1.4 | 27.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 1.1 | 20.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 1.0 | 8.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.8 | 25.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.8 | 20.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.8 | 9.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.7 | 10.7 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.6 | 9.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.6 | 15.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.6 | 6.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.6 | 17.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.6 | 6.8 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.6 | 15.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.5 | 11.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.5 | 8.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 10.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 5.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 8.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 2.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 6.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.3 | 17.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 10.7 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.3 | 5.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.3 | 5.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.3 | 3.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 3.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 7.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 4.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 6.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 2.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 10.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 16.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 14.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 4.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 2.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.2 | 9.8 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 4.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 29.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 3.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 4.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 8.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 5.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 3.5 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 7.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 6.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 3.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 4.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 2.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 12.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 6.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.9 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 2.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 4.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.4 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.1 | 3.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 5.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.6 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.1 | 6.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 3.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.9 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 1.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 2.3 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |