Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
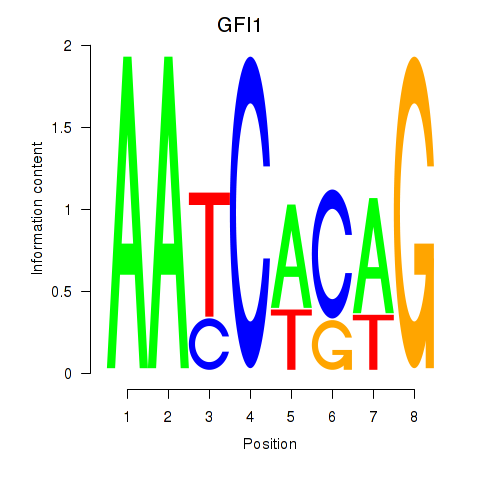
Results for GFI1
Z-value: 5.24
Transcription factors associated with GFI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GFI1
|
ENSG00000162676.12 | GFI1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GFI1 | hg38_v1_chr1_-_92483947_92483960, hg38_v1_chr1_-_92486916_92486932, hg38_v1_chr1_-_92486049_92486104 | 0.26 | 7.8e-05 | Click! |
Activity profile of GFI1 motif
Sorted Z-values of GFI1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GFI1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 2.5 | 9.9 | GO:0035548 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 2.5 | 7.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 2.4 | 16.6 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 2.2 | 6.5 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 2.1 | 6.3 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 1.9 | 7.8 | GO:0030821 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) |
| 1.8 | 9.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 1.8 | 5.4 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 1.8 | 5.3 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 1.7 | 32.1 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 1.7 | 6.6 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 1.6 | 18.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.6 | 11.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 1.6 | 3.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 1.6 | 4.7 | GO:0071988 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 1.5 | 6.1 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 1.4 | 12.9 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 1.3 | 3.8 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 1.2 | 10.0 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 1.2 | 3.5 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 1.2 | 6.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 1.0 | 4.1 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 1.0 | 2.9 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.9 | 6.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.9 | 2.8 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.9 | 3.6 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.9 | 6.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) cellular response to cobalt ion(GO:0071279) |
| 0.9 | 6.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.9 | 1.7 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.8 | 4.2 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.8 | 3.3 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.8 | 4.9 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.8 | 4.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.8 | 2.4 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.8 | 3.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.8 | 3.2 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.8 | 2.3 | GO:0061074 | regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.8 | 3.8 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.8 | 3.8 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.7 | 3.7 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.7 | 2.9 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.7 | 2.9 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.7 | 2.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.7 | 2.1 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.7 | 3.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.7 | 2.0 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.6 | 3.2 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.6 | 2.5 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.6 | 2.4 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.6 | 2.4 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.6 | 8.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.6 | 4.2 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.6 | 2.9 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.5 | 1.6 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.5 | 1.6 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.5 | 3.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.5 | 6.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.5 | 2.5 | GO:0035106 | negative regulation of luteinizing hormone secretion(GO:0033685) operant conditioning(GO:0035106) |
| 0.5 | 1.5 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.5 | 4.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.5 | 1.9 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 0.4 | 4.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.4 | 4.4 | GO:0009449 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.4 | 3.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.4 | 2.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 1.7 | GO:0045356 | toll-like receptor 7 signaling pathway(GO:0034154) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.4 | 3.7 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.4 | 6.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.4 | 2.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 3.5 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.4 | 3.9 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.4 | 2.2 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.4 | 1.9 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.4 | 2.6 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.4 | 3.7 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.4 | 3.7 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.4 | 0.4 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.4 | 5.4 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.3 | 14.0 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.3 | 10.8 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.3 | 13.0 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.3 | 3.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.3 | 5.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.3 | 3.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.3 | 0.7 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 0.3 | 2.0 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.3 | 2.3 | GO:0061085 | regulation of histone H3-K27 methylation(GO:0061085) positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.3 | 6.0 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.3 | 5.6 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.3 | 2.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.3 | 2.1 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.3 | 0.8 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.3 | 4.3 | GO:0021670 | lateral ventricle development(GO:0021670) nose development(GO:0043584) |
| 0.3 | 1.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.3 | 2.0 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.3 | 10.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.3 | 1.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.3 | 1.8 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.2 | 1.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.2 | 1.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.2 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.2 | 3.6 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.2 | 2.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 4.8 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 1.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 3.3 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.2 | 4.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 3.9 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 8.2 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 1.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 1.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 1.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 2.2 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.2 | 1.3 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.2 | 3.5 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 6.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 5.0 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.2 | 1.6 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.2 | 1.0 | GO:0070474 | tachykinin receptor signaling pathway(GO:0007217) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.2 | 0.8 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 7.8 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.2 | 2.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 1.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 8.8 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 7.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 4.0 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 3.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.2 | 17.3 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.2 | 7.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 1.6 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 1.4 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 | 4.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 1.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 3.3 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 12.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 2.0 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.2 | 0.5 | GO:0035963 | cellular response to interleukin-13(GO:0035963) |
| 0.2 | 3.4 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.2 | 8.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 8.0 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.1 | 8.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 5.8 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 2.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 3.5 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 5.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 3.6 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 5.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 2.5 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.1 | 2.8 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 3.7 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 4.5 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 3.0 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.1 | 16.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.2 | GO:0042269 | regulation of natural killer cell mediated cytotoxicity(GO:0042269) |
| 0.1 | 1.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 1.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 2.8 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 0.6 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 | 1.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 2.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 2.5 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 3.2 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 1.5 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 1.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 3.1 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.1 | 1.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 6.9 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 0.6 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 3.5 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 9.7 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.1 | 0.3 | GO:0072278 | thyroid-stimulating hormone secretion(GO:0070460) comma-shaped body morphogenesis(GO:0072049) metanephric comma-shaped body morphogenesis(GO:0072278) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.9 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 1.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 3.2 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 2.7 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 3.3 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 1.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 1.2 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.9 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 4.0 | GO:0008306 | associative learning(GO:0008306) |
| 0.1 | 3.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 14.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 2.9 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.1 | 3.9 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.1 | 0.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.2 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 2.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 1.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 3.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 4.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 15.9 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.8 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 1.5 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 1.5 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 1.4 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 1.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 7.7 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 1.2 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 3.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 1.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 5.3 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.9 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 2.9 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 0.7 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 2.2 | GO:0007163 | establishment or maintenance of cell polarity(GO:0007163) |
| 0.0 | 0.8 | GO:0017158 | regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.0 | 0.0 | GO:0071073 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 1.7 | 10.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 1.6 | 4.7 | GO:0055028 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 1.5 | 5.8 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 1.1 | 4.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 1.0 | 5.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.8 | 3.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.8 | 3.1 | GO:0000801 | central element(GO:0000801) |
| 0.7 | 4.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.7 | 2.1 | GO:0070701 | mucus layer(GO:0070701) |
| 0.7 | 4.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.6 | 3.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.6 | 1.8 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.6 | 4.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.6 | 4.6 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.6 | 6.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.5 | 3.6 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.5 | 1.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.5 | 8.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 5.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.4 | 6.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.4 | 1.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.4 | 9.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 2.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.3 | 1.7 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.3 | 8.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 2.5 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.3 | 3.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 2.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 3.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 3.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 2.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 12.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 3.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 4.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 2.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 2.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 3.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 9.7 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.2 | 3.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 12.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 9.5 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 1.7 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 5.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 1.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 20.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 0.5 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 6.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 4.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 34.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 4.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 3.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 5.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 3.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 10.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 4.3 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 1.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 17.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 2.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 10.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 8.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 14.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 2.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.0 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 1.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 5.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 18.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 4.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 8.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 4.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 5.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.9 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 4.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 119.9 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.0 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 2.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 12.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.7 | GO:0031966 | mitochondrial membrane(GO:0031966) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 32.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 2.5 | 9.9 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 2.3 | 9.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 2.1 | 6.3 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 2.0 | 8.2 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 2.0 | 6.1 | GO:0030305 | heparanase activity(GO:0030305) |
| 1.7 | 5.1 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 1.7 | 11.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.7 | 5.1 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 1.7 | 13.4 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 1.4 | 4.3 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 1.4 | 16.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 1.3 | 4.0 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 1.3 | 3.9 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 1.2 | 4.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.2 | 6.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 1.1 | 3.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.1 | 3.2 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 1.1 | 4.2 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.9 | 19.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.9 | 3.7 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.9 | 2.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.8 | 2.5 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.8 | 3.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.8 | 4.9 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.8 | 3.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.7 | 2.2 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.6 | 1.9 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.6 | 2.5 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.6 | 10.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.6 | 2.9 | GO:0070573 | tripeptidyl-peptidase activity(GO:0008240) peptidyl-dipeptidase activity(GO:0008241) bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 0.6 | 6.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.6 | 5.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.5 | 1.6 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.5 | 6.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.5 | 3.6 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.5 | 3.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.5 | 3.0 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.5 | 2.4 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.5 | 1.5 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.5 | 1.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.5 | 2.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.5 | 4.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.5 | 4.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.5 | 1.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.5 | 4.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.4 | 2.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.4 | 1.3 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.4 | 3.8 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.4 | 2.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.4 | 1.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.4 | 3.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.4 | 1.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.4 | 2.0 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.4 | 1.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.4 | 5.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.4 | 7.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.4 | 9.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 3.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 11.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 7.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 8.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.3 | 4.0 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.3 | 7.4 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.3 | 4.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 5.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 3.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.3 | 3.0 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.3 | 5.0 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.3 | 3.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.3 | 8.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 9.1 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 5.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 1.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.3 | 2.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 0.8 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.3 | 5.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.3 | 7.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 1.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 3.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 1.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 0.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 1.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 1.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.2 | 0.9 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 2.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 0.8 | GO:0086077 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.2 | 1.9 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 12.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 2.4 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.2 | 2.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 1.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.2 | 4.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 1.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 3.6 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 0.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 1.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.4 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 1.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 4.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 4.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 2.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 4.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 2.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 5.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 4.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 1.7 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 8.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 3.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 2.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.7 | GO:0004630 | heme oxygenase (decyclizing) activity(GO:0004392) phospholipase D activity(GO:0004630) |
| 0.1 | 12.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 3.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 6.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 6.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.7 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 2.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 3.5 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.1 | 1.0 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.3 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 1.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 2.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 4.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 9.5 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 1.1 | GO:0015250 | water transmembrane transporter activity(GO:0005372) water channel activity(GO:0015250) |
| 0.1 | 0.8 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 7.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 3.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 6.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 7.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 2.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 4.5 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 8.5 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 5.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 22.0 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 1.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 4.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 33.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 3.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.9 | GO:0071617 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 2.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 8.0 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 1.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.0 | 1.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 15.7 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 3.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 5.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.0 | GO:0051087 | chaperone binding(GO:0051087) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 17.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 10.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 3.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.2 | 3.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 3.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 20.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 5.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 5.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 5.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 6.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 8.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 3.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 7.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 2.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 4.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 6.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 4.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 17.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 10.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 6.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 16.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 5.3 | NABA MATRISOME | Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins |
| 0.0 | 0.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.7 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.5 | 3.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.5 | 4.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.5 | 9.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.5 | 7.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.4 | 5.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 7.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.4 | 28.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 7.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 5.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.4 | 7.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 16.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 8.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 9.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 12.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 0.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 6.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 4.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 4.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 7.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 5.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 2.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 4.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 6.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 7.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 3.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 3.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 5.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 2.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 1.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 13.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 3.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 5.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 3.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 5.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 1.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 4.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 14.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 8.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 5.0 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.1 | 6.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 5.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 22.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 4.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 2.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 3.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.2 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 2.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 0.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 5.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 1.0 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 3.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.2 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 0.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 4.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 5.3 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 8.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 8.3 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 3.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 0.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 2.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.3 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |