Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
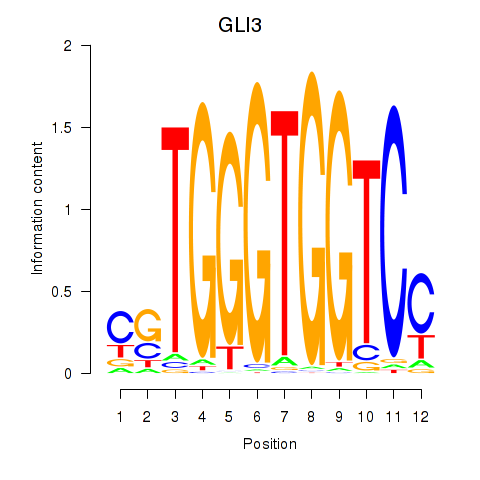
Results for GLI3
Z-value: 13.52
Transcription factors associated with GLI3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLI3
|
ENSG00000106571.15 | GLI3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI3 | hg38_v1_chr7_-_42152396_42152440, hg38_v1_chr7_-_42152444_42152458, hg38_v1_chr7_-_42237187_42237216 | 0.68 | 6.0e-31 | Click! |
Activity profile of GLI3 motif
Sorted Z-values of GLI3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GLI3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 25.7 | 102.9 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 18.1 | 90.7 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 11.6 | 34.8 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 10.0 | 60.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 9.5 | 28.4 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 7.9 | 55.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 6.8 | 47.9 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 6.1 | 18.2 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 6.0 | 18.1 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 5.8 | 28.9 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 5.4 | 21.4 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 5.2 | 15.6 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 5.2 | 31.0 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 5.0 | 15.0 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 5.0 | 14.9 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 4.9 | 24.4 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 4.7 | 14.1 | GO:0045957 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 4.6 | 18.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 4.5 | 13.5 | GO:0060032 | notochord regression(GO:0060032) |
| 4.4 | 21.9 | GO:0030070 | insulin processing(GO:0030070) |
| 4.3 | 38.8 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 4.3 | 21.4 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 4.2 | 25.2 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 4.2 | 16.7 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 4.1 | 12.4 | GO:0071812 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 4.1 | 12.3 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 4.1 | 20.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 3.8 | 11.3 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 3.7 | 11.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 3.6 | 10.7 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 3.4 | 20.5 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 3.3 | 10.0 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 3.3 | 13.3 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 3.3 | 42.7 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 3.2 | 9.5 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 3.1 | 24.5 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 3.0 | 9.0 | GO:0030221 | basophil differentiation(GO:0030221) positive regulation of chromatin assembly or disassembly(GO:0045799) astrocyte fate commitment(GO:0060018) |
| 3.0 | 41.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 2.9 | 14.7 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 2.9 | 11.5 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 2.9 | 2.9 | GO:0001554 | luteolysis(GO:0001554) |
| 2.8 | 28.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 2.8 | 8.4 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 2.7 | 16.4 | GO:0090625 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 2.6 | 5.2 | GO:0070256 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 2.5 | 43.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 2.5 | 69.0 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 2.5 | 14.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 2.5 | 29.5 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 2.4 | 12.2 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 2.4 | 16.9 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 2.4 | 7.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 2.4 | 7.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 2.3 | 7.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 2.3 | 25.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 2.3 | 11.5 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 2.3 | 11.4 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 2.2 | 6.5 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 2.1 | 10.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 2.1 | 6.4 | GO:0043605 | antibiotic metabolic process(GO:0016999) cellular amide catabolic process(GO:0043605) |
| 2.1 | 10.5 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 2.0 | 6.0 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 2.0 | 9.9 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 2.0 | 7.8 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.9 | 3.8 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 1.9 | 5.7 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 1.9 | 15.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 1.9 | 9.4 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 1.9 | 7.5 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.8 | 5.5 | GO:0046110 | purine nucleobase catabolic process(GO:0006145) xanthine metabolic process(GO:0046110) |
| 1.8 | 12.8 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 1.8 | 5.4 | GO:0001757 | somite specification(GO:0001757) |
| 1.8 | 5.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 1.8 | 14.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 1.8 | 7.0 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 1.7 | 12.2 | GO:0015705 | iodide transport(GO:0015705) |
| 1.7 | 5.2 | GO:0050748 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) negative regulation of lipoprotein metabolic process(GO:0050748) |
| 1.7 | 10.4 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 1.7 | 10.3 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 1.7 | 3.4 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 1.7 | 5.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 1.7 | 11.8 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 1.7 | 11.7 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 1.7 | 5.0 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 1.7 | 24.8 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 1.6 | 8.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 1.6 | 4.9 | GO:0007525 | somatic muscle development(GO:0007525) |
| 1.6 | 11.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 1.6 | 4.8 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 1.6 | 8.0 | GO:0072014 | proximal tubule development(GO:0072014) |
| 1.6 | 6.4 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 1.6 | 11.0 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 1.6 | 14.1 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 1.6 | 6.2 | GO:1903633 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 1.6 | 6.2 | GO:1904141 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 1.5 | 9.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.5 | 7.7 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 1.5 | 4.5 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 1.5 | 72.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 1.5 | 7.4 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 1.5 | 13.3 | GO:0007135 | meiosis II(GO:0007135) |
| 1.4 | 8.7 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 1.4 | 11.3 | GO:0006477 | protein sulfation(GO:0006477) |
| 1.4 | 4.2 | GO:1902996 | regulation of neurofibrillary tangle assembly(GO:1902996) |
| 1.4 | 12.7 | GO:0009249 | protein lipoylation(GO:0009249) |
| 1.4 | 23.9 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.4 | 4.2 | GO:0071486 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 1.4 | 4.1 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 1.4 | 12.3 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 1.4 | 16.4 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 1.4 | 17.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 1.4 | 12.2 | GO:0046959 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 1.3 | 76.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 1.3 | 5.4 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 1.3 | 3.9 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 1.3 | 17.0 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 1.3 | 12.7 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 1.3 | 11.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 1.2 | 6.2 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 1.2 | 28.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 1.2 | 4.9 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.2 | 8.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 1.2 | 4.7 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 1.1 | 11.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 1.1 | 6.8 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 1.1 | 5.6 | GO:2000834 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 1.1 | 7.9 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 1.1 | 6.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 1.1 | 2.1 | GO:0002351 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 1.1 | 3.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 1.1 | 19.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 1.0 | 14.6 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 1.0 | 3.1 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 1.0 | 11.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 1.0 | 7.0 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 1.0 | 4.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.0 | 3.0 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 1.0 | 4.0 | GO:0032571 | response to vitamin K(GO:0032571) response to hydroxyisoflavone(GO:0033594) |
| 1.0 | 4.0 | GO:0010157 | response to chlorate(GO:0010157) |
| 1.0 | 12.8 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 1.0 | 1.0 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 1.0 | 2.9 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 1.0 | 13.6 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 1.0 | 32.7 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 1.0 | 8.6 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 1.0 | 1.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 1.0 | 21.9 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.9 | 11.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.9 | 14.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.9 | 6.0 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.9 | 8.6 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.9 | 6.8 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.9 | 2.6 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.8 | 4.2 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.8 | 6.6 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.8 | 6.5 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.8 | 11.4 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.8 | 16.9 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.8 | 4.8 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.8 | 4.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.8 | 6.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.8 | 4.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.8 | 5.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.8 | 2.3 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.8 | 10.0 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.8 | 1.5 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.8 | 4.5 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.8 | 0.8 | GO:0072054 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.7 | 2.2 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.7 | 2.9 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.7 | 4.9 | GO:0045425 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.7 | 8.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.7 | 7.6 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.7 | 4.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.7 | 6.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.7 | 6.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.7 | 4.7 | GO:0097267 | lipid hydroxylation(GO:0002933) omega-hydroxylase P450 pathway(GO:0097267) |
| 0.7 | 16.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.7 | 6.8 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.7 | 4.7 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.7 | 8.0 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.7 | 5.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.7 | 9.9 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.7 | 11.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.7 | 36.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.7 | 2.0 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.6 | 3.9 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.6 | 5.7 | GO:1902255 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.6 | 10.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.6 | 1.9 | GO:1903892 | negative regulation of ATF6-mediated unfolded protein response(GO:1903892) |
| 0.6 | 10.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.6 | 34.7 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.6 | 14.2 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.6 | 1.8 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.6 | 17.4 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.6 | 2.3 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.6 | 8.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.6 | 2.8 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.6 | 12.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.6 | 8.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.6 | 12.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.6 | 10.0 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.6 | 3.3 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.6 | 5.0 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.6 | 3.9 | GO:0046618 | drug export(GO:0046618) |
| 0.6 | 3.3 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.6 | 6.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.6 | 2.2 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.5 | 4.3 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.5 | 8.6 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.5 | 3.2 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.5 | 9.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.5 | 1.0 | GO:0035419 | activation of MAPK activity involved in innate immune response(GO:0035419) |
| 0.5 | 5.6 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.5 | 1.0 | GO:0048541 | mucosal-associated lymphoid tissue development(GO:0048537) Peyer's patch development(GO:0048541) Peyer's patch morphogenesis(GO:0061146) |
| 0.5 | 8.0 | GO:0009068 | aspartate family amino acid catabolic process(GO:0009068) |
| 0.5 | 20.9 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.5 | 2.5 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.5 | 3.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.5 | 12.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.5 | 9.3 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.5 | 3.3 | GO:1900122 | positive regulation of keratinocyte migration(GO:0051549) positive regulation of receptor binding(GO:1900122) |
| 0.5 | 5.9 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.4 | 5.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.4 | 5.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.4 | 1.8 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.4 | 5.2 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.4 | 14.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.4 | 1.7 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.4 | 0.8 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.4 | 2.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.4 | 1.6 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.4 | 3.7 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.4 | 15.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.4 | 6.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.4 | 2.4 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.4 | 5.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.4 | 8.4 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.4 | 8.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.4 | 6.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.4 | 8.7 | GO:0007616 | long-term memory(GO:0007616) |
| 0.4 | 3.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.4 | 3.5 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.4 | 3.8 | GO:0072540 | T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.4 | 1.9 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.4 | 11.5 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.4 | 2.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.4 | 5.6 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.4 | 30.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.4 | 2.5 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.4 | 0.7 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.3 | 3.8 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.3 | 2.8 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.3 | 13.7 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.3 | 5.0 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.3 | 15.8 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.3 | 4.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 1.0 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.3 | 2.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.3 | 3.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 11.3 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 3.4 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 7.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.3 | 6.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.3 | 3.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.3 | 3.9 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 0.9 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.3 | 6.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 9.5 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.3 | 5.3 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.3 | 4.4 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.3 | 5.9 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.3 | 5.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 1.2 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.3 | 3.5 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.3 | 7.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.3 | 17.2 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.3 | 3.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 7.3 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.3 | 7.7 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.3 | 2.9 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.3 | 5.1 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.3 | 2.6 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.3 | 3.3 | GO:0050927 | positive regulation of positive chemotaxis(GO:0050927) |
| 0.3 | 1.8 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 1.5 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.2 | 0.7 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.2 | 3.2 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.2 | 4.2 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 1.3 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.2 | 2.4 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 1.7 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.2 | 2.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.2 | 1.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 2.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.2 | 4.5 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.2 | 1.6 | GO:0060453 | regulation of gastric acid secretion(GO:0060453) |
| 0.2 | 7.6 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.2 | 6.5 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.2 | 5.9 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.2 | 5.6 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.2 | 1.5 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.2 | 2.6 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.2 | 5.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 0.6 | GO:1900081 | negative regulation of cardiac muscle adaptation(GO:0010616) regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) negative regulation of adrenergic receptor signaling pathway(GO:0071878) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) negative regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901205) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.2 | 5.7 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 18.3 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.2 | 2.3 | GO:0007512 | adult heart development(GO:0007512) |
| 0.2 | 0.9 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.2 | 2.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.2 | 13.6 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.2 | 3.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.2 | 1.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 2.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 7.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 4.8 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.2 | 109.5 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.2 | 0.9 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 10.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.7 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.1 | 6.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.3 | GO:0010954 | positive regulation of protein processing(GO:0010954) |
| 0.1 | 1.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 6.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 3.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 15.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 2.2 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 2.9 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 8.5 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 1.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 9.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 2.4 | GO:0045821 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.1 | 5.6 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 5.2 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 3.7 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 1.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 1.8 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 1.4 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.1 | 1.4 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.1 | 3.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 1.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 8.6 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.1 | 1.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.9 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) cell differentiation involved in salivary gland development(GO:0060689) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.7 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 1.3 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.2 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) positive regulation of gastrulation(GO:2000543) |
| 0.1 | 0.5 | GO:0038007 | anterior/posterior axon guidance(GO:0033564) netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.6 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.1 | 3.9 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 1.9 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.1 | 0.9 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.2 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 0.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 1.6 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.1 | 5.2 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.1 | 6.4 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 0.8 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 1.0 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 2.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.9 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 1.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 2.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 2.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.6 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 1.1 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 5.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.0 | GO:0060711 | labyrinthine layer development(GO:0060711) |
| 0.0 | 0.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 1.0 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 1.2 | GO:0003279 | cardiac septum development(GO:0003279) |
| 0.0 | 0.8 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 1.0 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 1.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.4 | 37.7 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 6.9 | 111.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 5.1 | 10.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 5.1 | 15.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 4.6 | 22.8 | GO:0001652 | granular component(GO:0001652) |
| 4.4 | 70.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 4.1 | 102.8 | GO:0042627 | chylomicron(GO:0042627) |
| 4.0 | 20.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 3.7 | 44.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 3.6 | 21.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 3.2 | 9.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 3.1 | 15.5 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 3.0 | 8.9 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 3.0 | 17.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 2.9 | 71.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 2.7 | 8.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 2.5 | 20.4 | GO:0033648 | host cell cytoplasm(GO:0030430) host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) host cell cytoplasm part(GO:0033655) |
| 2.3 | 11.5 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 2.2 | 8.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 2.1 | 8.5 | GO:0070695 | FHF complex(GO:0070695) |
| 2.1 | 14.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 2.0 | 10.0 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 2.0 | 18.0 | GO:0030897 | HOPS complex(GO:0030897) |
| 2.0 | 29.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 2.0 | 11.7 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 1.8 | 29.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 1.8 | 15.9 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 1.7 | 5.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 1.6 | 16.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 1.6 | 7.9 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 1.6 | 7.8 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 1.5 | 47.9 | GO:0034706 | sodium channel complex(GO:0034706) |
| 1.5 | 6.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 1.5 | 7.7 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 1.5 | 11.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 1.4 | 11.5 | GO:0045179 | apical cortex(GO:0045179) |
| 1.4 | 4.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 1.3 | 4.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 1.3 | 9.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 1.3 | 14.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 1.3 | 89.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 1.3 | 7.5 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 1.2 | 11.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.2 | 8.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 1.2 | 40.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 1.2 | 21.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 1.1 | 20.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.1 | 3.4 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 1.1 | 5.5 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 1.0 | 6.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 1.0 | 6.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 1.0 | 3.0 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 1.0 | 3.8 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.9 | 8.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.9 | 10.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.9 | 15.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.9 | 6.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.9 | 11.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.9 | 4.3 | GO:0044308 | axonal spine(GO:0044308) |
| 0.9 | 34.2 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.8 | 4.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.8 | 14.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.8 | 16.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.8 | 12.3 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.8 | 9.9 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.8 | 6.0 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.7 | 5.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.7 | 5.9 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.7 | 5.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.7 | 2.9 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.7 | 8.7 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.6 | 6.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.6 | 1.9 | GO:0043293 | apoptosome(GO:0043293) |
| 0.6 | 8.1 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.6 | 11.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.6 | 6.5 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.6 | 18.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.6 | 18.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.5 | 3.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.5 | 1.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.5 | 9.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.5 | 14.0 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.5 | 3.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.5 | 6.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.5 | 7.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.5 | 3.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.5 | 3.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.5 | 12.5 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.5 | 24.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.5 | 16.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.4 | 12.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.4 | 12.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.4 | 15.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.4 | 5.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.4 | 23.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.4 | 11.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.4 | 26.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.4 | 6.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.3 | 13.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.3 | 3.0 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.3 | 4.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 2.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.3 | 12.5 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.3 | 7.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.3 | 1.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.3 | 7.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.3 | 18.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.3 | 13.5 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.3 | 11.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.3 | 13.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 6.2 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 0.7 | GO:0033593 | BRCA2-MAGE-D1 complex(GO:0033593) |
| 0.2 | 4.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.2 | 1.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 3.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 0.7 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 2.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 7.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 7.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 0.8 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.2 | 2.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 4.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 15.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 6.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 53.3 | GO:0001726 | ruffle(GO:0001726) |
| 0.2 | 18.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 3.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.2 | 45.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 3.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 4.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 21.6 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 20.3 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 10.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 3.8 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 3.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 126.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 5.8 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 3.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 3.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 9.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 7.5 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 4.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 7.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 1.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 18.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 6.5 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 0.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 8.6 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 0.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 6.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 6.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 14.4 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 6.5 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.1 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 11.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.4 | 102.8 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 10.0 | 60.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 9.4 | 37.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 7.7 | 38.3 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 5.7 | 28.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 5.0 | 34.8 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 4.7 | 14.1 | GO:0004877 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 4.4 | 21.8 | GO:0050610 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 4.3 | 55.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 4.2 | 16.8 | GO:0005334 | norepinephrine:sodium symporter activity(GO:0005334) |
| 3.9 | 90.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 3.9 | 35.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 3.6 | 21.4 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 3.4 | 10.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 3.4 | 16.9 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 3.2 | 9.5 | GO:0032093 | SAM domain binding(GO:0032093) |
| 3.1 | 9.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 3.0 | 23.8 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 3.0 | 11.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 2.9 | 8.7 | GO:0070538 | oleic acid binding(GO:0070538) |
| 2.8 | 33.7 | GO:0008430 | selenium binding(GO:0008430) |
| 2.8 | 56.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 2.7 | 16.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 2.7 | 13.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 2.7 | 10.6 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 2.6 | 31.0 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 2.5 | 10.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 2.4 | 22.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 2.4 | 7.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 2.3 | 18.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 2.1 | 8.6 | GO:0005497 | androgen binding(GO:0005497) |
| 2.1 | 12.8 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 2.1 | 62.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 2.1 | 6.2 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 2.1 | 6.2 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 2.0 | 28.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 2.0 | 8.1 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 2.0 | 8.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.9 | 5.8 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.9 | 5.8 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 1.9 | 5.7 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 1.9 | 15.0 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 1.9 | 9.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 1.8 | 5.5 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 1.8 | 9.0 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) apolipoprotein A-I receptor binding(GO:0034191) |
| 1.8 | 21.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.8 | 24.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 1.7 | 28.6 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 1.6 | 14.7 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 1.6 | 11.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 1.6 | 25.8 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 1.6 | 6.4 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 1.6 | 26.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.6 | 6.2 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 1.5 | 10.7 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 1.5 | 7.6 | GO:0004803 | transposase activity(GO:0004803) |
| 1.5 | 6.0 | GO:0031716 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 1.5 | 10.5 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 1.5 | 13.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 1.4 | 10.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 1.4 | 10.0 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 1.4 | 5.7 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 1.4 | 5.6 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 1.4 | 8.4 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 1.4 | 4.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 1.4 | 8.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 1.4 | 16.7 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 1.4 | 5.5 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 1.3 | 4.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 1.3 | 17.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 1.3 | 5.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.3 | 5.0 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.2 | 6.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.2 | 13.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 1.2 | 17.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 1.2 | 2.4 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.2 | 2.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 1.2 | 21.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 1.1 | 8.0 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 1.1 | 15.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 1.1 | 14.6 | GO:0019841 | retinol binding(GO:0019841) |
| 1.1 | 5.6 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 1.1 | 7.8 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 1.1 | 66.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 1.1 | 8.7 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 1.1 | 22.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 1.0 | 5.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 1.0 | 6.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 1.0 | 3.0 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 1.0 | 4.0 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 1.0 | 14.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 1.0 | 3.9 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 1.0 | 8.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 1.0 | 8.6 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.9 | 27.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.9 | 5.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.9 | 16.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.9 | 11.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.9 | 12.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.9 | 14.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.9 | 18.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.8 | 8.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.8 | 2.5 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.8 | 6.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.8 | 1.6 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.8 | 4.0 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.8 | 11.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.8 | 2.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.8 | 6.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.8 | 6.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.8 | 12.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.8 | 5.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.7 | 3.6 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.7 | 8.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.7 | 2.8 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.7 | 6.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.7 | 11.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.7 | 18.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.7 | 4.8 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.7 | 2.7 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.7 | 13.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.7 | 16.9 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.6 | 3.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.6 | 14.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.6 | 13.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.6 | 9.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.6 | 5.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.6 | 11.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.6 | 20.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.6 | 2.4 | GO:0019862 | IgA binding(GO:0019862) |
| 0.6 | 3.5 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.6 | 4.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.6 | 2.3 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.6 | 9.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.6 | 9.6 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.6 | 2.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.6 | 18.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.6 | 9.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.6 | 3.9 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.5 | 7.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.5 | 15.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.5 | 9.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.5 | 5.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.5 | 3.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.5 | 3.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.5 | 4.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 20.9 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.5 | 3.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.5 | 59.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.5 | 2.9 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.5 | 1.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.5 | 7.9 | GO:0019864 | IgG binding(GO:0019864) |
| 0.5 | 9.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.5 | 11.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.5 | 31.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.4 | 8.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.4 | 3.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.4 | 2.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.4 | 12.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.4 | 6.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 8.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.4 | 5.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 4.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.4 | 24.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.4 | 2.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.4 | 2.0 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.4 | 2.4 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.4 | 4.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.4 | 12.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.4 | 2.6 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.4 | 1.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.4 | 6.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.4 | 2.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 4.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.4 | 6.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.4 | 8.9 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.4 | 8.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.4 | 8.5 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.3 | 15.7 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.3 | 1.4 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.3 | 6.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.3 | 3.7 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.3 | 11.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 16.3 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.3 | 20.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 4.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.3 | 2.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 3.4 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.3 | 5.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.3 | 8.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.3 | 51.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.3 | 5.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.3 | 7.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 19.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.3 | 25.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 5.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.3 | 8.8 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.3 | 7.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.3 | 4.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 7.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 1.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 2.9 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.3 | 3.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 7.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 1.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.2 | 2.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 25.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 8.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 2.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 25.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.2 | 29.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.2 | 2.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 6.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.2 | 18.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 3.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 0.8 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.2 | 6.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 4.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 5.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 1.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 1.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.2 | 7.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 18.4 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.2 | 4.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 1.3 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.2 | 0.7 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.2 | 6.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 3.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 1.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.2 | 2.0 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 5.6 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.1 | 0.7 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 5.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 6.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 9.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 33.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 5.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 1.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 7.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 3.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 4.6 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 1.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 3.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 2.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 4.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 4.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 2.2 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 7.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.5 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 17.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 17.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 7.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 10.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 0.7 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 3.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 3.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 4.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 1.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.7 | GO:0070008 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 11.7 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 7.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 29.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 1.1 | 54.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.8 | 110.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.7 | 38.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.7 | 8.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.7 | 30.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.7 | 10.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.7 | 7.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.6 | 27.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.6 | 27.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.6 | 6.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.6 | 14.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.6 | 14.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.6 | 22.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.5 | 35.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.5 | 22.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.5 | 10.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.5 | 24.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.5 | 8.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.4 | 16.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.4 | 6.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.4 | 14.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.4 | 4.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.4 | 5.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.4 | 19.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.4 | 8.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.4 | 6.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 5.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.4 | 17.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.3 | 7.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.3 | 7.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 11.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 5.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 7.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 5.6 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.3 | 8.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 4.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.3 | 11.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.3 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 9.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.2 | 9.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 3.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 3.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.2 | 3.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 9.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.2 | 6.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 5.6 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 15.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 2.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 7.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 4.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 36.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 11.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 1.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 6.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 6.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 40.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 5.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.9 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 1.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 1.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 3.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 1.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 2.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 7.1 | REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | Genes involved in Downstream signaling of activated FGFR |
| 2.7 | 67.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 2.2 | 43.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 2.1 | 12.7 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 2.0 | 4.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 1.4 | 62.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.3 | 52.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 1.3 | 21.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 1.3 | 35.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 1.3 | 28.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.2 | 24.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 1.1 | 16.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 1.0 | 25.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 1.0 | 6.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 1.0 | 18.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.9 | 17.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.9 | 70.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.9 | 34.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.9 | 10.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.9 | 24.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.7 | 14.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.6 | 28.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.6 | 14.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.6 | 17.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.6 | 12.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.6 | 8.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.6 | 13.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.5 | 6.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.5 | 11.9 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.5 | 6.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.5 | 10.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.5 | 32.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.5 | 8.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.5 | 11.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.5 | 45.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.5 | 6.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.5 | 4.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.5 | 1.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.4 | 7.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.4 | 7.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 25.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.4 | 15.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.4 | 7.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 53.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.4 | 15.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.4 | 9.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.4 | 8.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.4 | 6.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.4 | 7.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 4.8 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.4 | 10.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 4.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 66.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.3 | 4.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.3 | 4.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 11.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 4.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 22.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.3 | 7.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 11.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.3 | 3.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 6.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.3 | 13.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 8.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.3 | 8.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.3 | 7.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 6.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.3 | 35.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 9.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 23.0 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.2 | 14.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 6.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 2.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 2.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 4.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.2 | 7.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 5.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 5.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 4.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 2.0 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.2 | 7.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 5.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 13.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 2.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 15.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 8.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 14.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 7.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 1.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 7.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 27.8 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 2.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 4.6 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.7 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 5.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 3.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |