Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
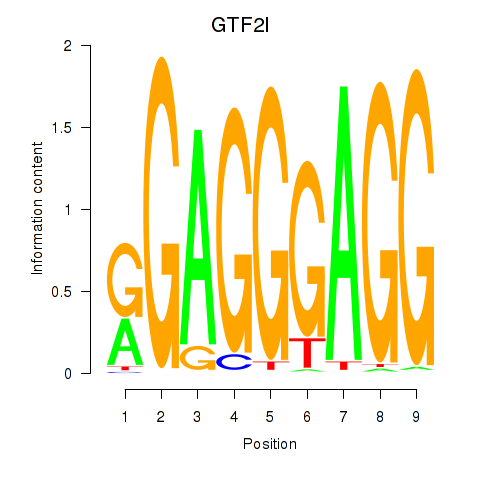
Results for GTF2I
Z-value: 2.38
Transcription factors associated with GTF2I
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GTF2I
|
ENSG00000263001.7 | GTF2I |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GTF2I | hg38_v1_chr7_+_74657695_74657743, hg38_v1_chr7_+_74650224_74650240 | 0.47 | 2.1e-13 | Click! |
Activity profile of GTF2I motif
Sorted Z-values of GTF2I motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GTF2I
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 17.9 | GO:0090176 | microtubule cytoskeleton organization involved in establishment of planar polarity(GO:0090176) |
| 3.7 | 18.5 | GO:0006344 | optic cup formation involved in camera-type eye development(GO:0003408) maintenance of chromatin silencing(GO:0006344) |
| 3.4 | 10.2 | GO:0021897 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 3.0 | 9.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 3.0 | 14.9 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 2.9 | 8.8 | GO:0002368 | B cell cytokine production(GO:0002368) |
| 2.7 | 27.3 | GO:0060136 | enucleate erythrocyte differentiation(GO:0043353) embryonic process involved in female pregnancy(GO:0060136) |
| 2.7 | 10.8 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) negative regulation of glial cell migration(GO:1903976) |
| 2.5 | 7.4 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 2.4 | 28.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 2.3 | 25.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 2.2 | 11.0 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 2.1 | 14.7 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 2.1 | 6.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 2.0 | 6.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.9 | 13.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.8 | 23.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 1.8 | 3.6 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 1.8 | 8.9 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 1.7 | 19.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 1.7 | 26.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 1.6 | 4.7 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 1.5 | 10.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 1.5 | 6.0 | GO:1904021 | negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of G-protein coupled receptor internalization(GO:1904020) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 1.4 | 4.3 | GO:1902728 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 1.4 | 4.2 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 1.4 | 12.6 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 1.4 | 5.4 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 1.3 | 5.4 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 1.3 | 5.4 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 1.2 | 4.9 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 1.2 | 4.8 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 1.2 | 3.6 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 1.2 | 4.7 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 1.2 | 3.5 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.2 | 5.9 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 1.2 | 12.7 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.1 | 3.3 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 1.1 | 4.4 | GO:0036483 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 1.1 | 4.3 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 1.1 | 7.5 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 1.0 | 9.8 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.0 | 22.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 1.0 | 1.9 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.9 | 5.6 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.9 | 3.7 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.9 | 3.6 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.9 | 5.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.9 | 7.0 | GO:0033484 | regulation of protein ADP-ribosylation(GO:0010835) nitric oxide homeostasis(GO:0033484) |
| 0.9 | 2.6 | GO:0060957 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 0.9 | 1.7 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.9 | 3.4 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.8 | 4.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.8 | 3.4 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.8 | 4.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.8 | 4.0 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.8 | 5.5 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.8 | 7.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.8 | 7.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.8 | 5.5 | GO:1902473 | regulation of protein localization to synapse(GO:1902473) |
| 0.8 | 11.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.8 | 3.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.8 | 4.6 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.8 | 5.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.8 | 5.3 | GO:1905123 | regulation of glucosylceramidase activity(GO:1905123) |
| 0.7 | 2.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.7 | 7.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.7 | 2.2 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.7 | 1.5 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.7 | 6.6 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.7 | 2.2 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.7 | 2.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.7 | 2.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.7 | 2.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.7 | 10.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.7 | 2.1 | GO:1901656 | glucoside transport(GO:0042946) glycoside transport(GO:1901656) |
| 0.7 | 6.9 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.7 | 2.7 | GO:0048627 | myoblast development(GO:0048627) |
| 0.7 | 10.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.7 | 2.0 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.6 | 5.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.6 | 1.9 | GO:0071314 | cellular response to cocaine(GO:0071314) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.6 | 3.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.6 | 8.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.6 | 2.5 | GO:0060926 | cardiac pacemaker cell differentiation(GO:0060920) cardiac pacemaker cell development(GO:0060926) |
| 0.6 | 8.7 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.6 | 3.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.6 | 19.5 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.6 | 7.0 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.6 | 11.6 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.6 | 1.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.6 | 1.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.5 | 3.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.5 | 14.1 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.5 | 4.9 | GO:0060613 | fat pad development(GO:0060613) |
| 0.5 | 4.8 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.5 | 1.6 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.5 | 2.6 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.5 | 4.6 | GO:1990668 | vesicle fusion with endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membrane(GO:1990668) |
| 0.5 | 0.5 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.5 | 1.5 | GO:0060382 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.5 | 1.5 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.5 | 4.0 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.5 | 2.0 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.5 | 4.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.5 | 1.9 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.5 | 3.9 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.5 | 6.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.5 | 2.9 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.5 | 1.4 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.5 | 3.3 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.5 | 2.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.5 | 2.3 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.5 | 1.4 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.5 | 5.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.5 | 1.4 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.5 | 4.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.5 | 1.4 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.4 | 1.3 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.4 | 6.7 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.4 | 3.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.4 | 6.7 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.4 | 41.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.4 | 9.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.4 | 0.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.4 | 12.6 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.4 | 12.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.4 | 1.7 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.4 | 5.1 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.4 | 1.3 | GO:0055020 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.4 | 3.8 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.4 | 1.7 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.4 | 2.5 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.4 | 8.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.4 | 2.9 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.4 | 4.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.4 | 2.1 | GO:1903275 | regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.4 | 10.7 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.4 | 4.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.4 | 2.4 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.4 | 4.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 4.8 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.4 | 1.2 | GO:1901805 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.4 | 15.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.4 | 3.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.4 | 0.8 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.4 | 0.4 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.4 | 4.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.4 | 0.4 | GO:0061443 | endocardial cushion cell differentiation(GO:0061443) |
| 0.4 | 1.9 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.4 | 3.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 7.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.4 | 3.3 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.4 | 1.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) negative regulation of lymphangiogenesis(GO:1901491) |
| 0.4 | 5.8 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.4 | 2.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.4 | 2.5 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.4 | 1.8 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.4 | 1.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.4 | 1.4 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.3 | 1.7 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.3 | 0.7 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.3 | 4.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.3 | 2.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.3 | 6.4 | GO:0035646 | endosome to melanosome transport(GO:0035646) pigment accumulation(GO:0043476) cellular pigment accumulation(GO:0043482) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.3 | 2.4 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.3 | 1.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.3 | 4.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 1.3 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.3 | 1.3 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) peptidyl-glycine modification(GO:0018201) |
| 0.3 | 0.7 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.3 | 4.2 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.3 | 2.6 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 0.6 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.3 | 2.2 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.3 | 2.9 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.3 | 0.9 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.3 | 2.8 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.3 | 1.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.3 | 1.2 | GO:2000077 | negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.3 | 3.7 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.3 | 2.1 | GO:0001712 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.3 | 1.8 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.3 | 4.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.3 | 2.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.3 | 1.5 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 5.3 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.3 | 1.8 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.3 | 2.0 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.3 | 1.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 1.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.3 | 3.4 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.3 | 4.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 2.3 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.3 | 2.3 | GO:0007379 | segment specification(GO:0007379) |
| 0.3 | 1.7 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 4.8 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.3 | 1.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.3 | 5.2 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.3 | 1.6 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.3 | 1.1 | GO:0061360 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.3 | 1.9 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.3 | 5.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.3 | 1.6 | GO:0060356 | leucine import(GO:0060356) |
| 0.3 | 1.6 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 0.8 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.3 | 1.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 | 16.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.3 | 1.8 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.3 | 2.5 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.3 | 2.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 5.2 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.2 | 0.7 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.2 | 5.9 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.2 | 0.7 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 | 7.1 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.2 | 4.6 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.2 | 1.0 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.2 | 6.0 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.2 | 6.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 3.5 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.2 | 0.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 0.9 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.2 | 2.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 0.5 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.2 | 1.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.2 | 1.3 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 0.2 | 2.8 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.2 | 4.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 0.6 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.2 | 0.6 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.2 | 1.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 1.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 4.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 1.7 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.2 | 5.4 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.2 | 0.6 | GO:0072183 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.2 | 3.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.2 | 1.4 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 1.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 1.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 0.2 | GO:0043366 | beta selection(GO:0043366) |
| 0.2 | 3.0 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.2 | 0.8 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 1.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 3.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 9.8 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.2 | 1.6 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.2 | 9.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 1.2 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.2 | 2.9 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 1.7 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.2 | 0.8 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.2 | 4.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 1.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.2 | 1.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.2 | 2.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 2.5 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.2 | 1.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 7.9 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.2 | 7.6 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.2 | 0.5 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.2 | 2.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 1.2 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.2 | 2.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 0.9 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.2 | 1.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 0.5 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.2 | 0.7 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.2 | 2.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 2.0 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.2 | 1.2 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 5.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 0.6 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.2 | 0.5 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 | 0.9 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.2 | 2.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.2 | 11.6 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.2 | 13.2 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.2 | 2.6 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.2 | 1.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 1.7 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.2 | 3.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.2 | 0.5 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 3.5 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.1 | 2.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.6 | GO:2000255 | regulation of male germ cell proliferation(GO:2000254) negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 1.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 1.0 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.6 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 1.9 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.1 | 0.7 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 3.4 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 0.4 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.4 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 1.6 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 5.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 0.4 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 3.0 | GO:0031958 | corticosteroid receptor signaling pathway(GO:0031958) glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.1 | 0.5 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.1 | 0.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 4.5 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 0.6 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 1.3 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.5 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 9.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 2.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 4.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 3.4 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.1 | 0.8 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 2.1 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 1.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.9 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 0.2 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.1 | 1.0 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 3.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 2.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 2.4 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 4.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 3.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 2.0 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 1.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 1.4 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 0.7 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 1.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 0.6 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 | 1.9 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.3 | GO:2000617 | positive regulation of histone H3-K9 acetylation(GO:2000617) |
| 0.1 | 2.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.2 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 4.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 3.0 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.1 | 0.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 3.0 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 1.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 1.8 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 1.5 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 0.7 | GO:0055062 | phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 5.1 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 1.8 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 0.2 | GO:1903421 | regulation of synaptic vesicle recycling(GO:1903421) |
| 0.1 | 0.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 2.0 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 1.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.5 | GO:0051665 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) membrane raft localization(GO:0051665) |
| 0.1 | 1.0 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.7 | GO:0036344 | platelet morphogenesis(GO:0036344) |
| 0.1 | 5.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.4 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 1.1 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.1 | 0.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 2.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.8 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 3.5 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.1 | 2.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.1 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 0.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 1.9 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.2 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 0.1 | 0.3 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.1 | 2.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 5.8 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 2.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.7 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 1.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.5 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 0.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.1 | 2.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 1.0 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 4.4 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.1 | 4.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 1.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 2.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 0.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.4 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 1.2 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.6 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.3 | GO:1990564 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 1.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 1.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 1.3 | GO:1990090 | cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 3.3 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.2 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.5 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.3 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 1.2 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.9 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 1.6 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 1.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.3 | GO:2000774 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 1.2 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 1.5 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.3 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 4.9 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.2 | GO:0048296 | regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.6 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.8 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.8 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.7 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.4 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.8 | GO:1902583 | intracellular transport of virus(GO:0075733) multi-organism intracellular transport(GO:1902583) |
| 0.0 | 0.4 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 1.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.0 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.3 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 1.9 | 29.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 1.8 | 17.9 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 1.6 | 6.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 1.5 | 13.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 1.4 | 17.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 1.3 | 6.3 | GO:0031523 | Myb complex(GO:0031523) |
| 1.2 | 10.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 1.1 | 4.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.1 | 2.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 1.0 | 13.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 1.0 | 5.2 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.0 | 6.7 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.9 | 3.8 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.9 | 2.6 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.9 | 7.7 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.8 | 3.4 | GO:0071942 | XPC complex(GO:0071942) |
| 0.8 | 20.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.8 | 5.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.8 | 4.8 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.8 | 7.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.8 | 2.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.8 | 4.6 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.7 | 9.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.7 | 11.7 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.7 | 2.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.7 | 8.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.7 | 5.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.7 | 8.3 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.6 | 4.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.6 | 9.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.6 | 20.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.6 | 4.7 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.6 | 2.9 | GO:0031673 | H zone(GO:0031673) |
| 0.6 | 1.7 | GO:0043260 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.6 | 5.0 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.5 | 3.8 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.5 | 11.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.5 | 2.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.5 | 3.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.5 | 8.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 5.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.5 | 7.8 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.5 | 2.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.5 | 3.2 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.5 | 6.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.5 | 1.4 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.4 | 8.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.4 | 20.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.4 | 2.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 3.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.4 | 4.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.4 | 2.0 | GO:0044094 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.4 | 2.7 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.4 | 2.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 1.9 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 6.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.4 | 10.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.4 | 1.4 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.4 | 5.7 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.4 | 2.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.4 | 25.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 1.7 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.3 | 1.7 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.3 | 9.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.3 | 1.9 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.3 | 3.2 | GO:0051286 | cell tip(GO:0051286) |
| 0.3 | 5.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 2.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.3 | 4.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 3.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 5.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.3 | 9.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 8.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.3 | 2.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.3 | 11.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 3.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 5.0 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.3 | 1.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.3 | 16.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 1.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 2.6 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.3 | 1.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.3 | 2.8 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.3 | 2.5 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.2 | 52.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.2 | 1.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.2 | 2.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 5.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 1.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 3.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 1.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 2.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 2.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 2.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 4.2 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.2 | 1.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.2 | 14.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 3.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 29.1 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 4.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 1.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 2.8 | GO:0032059 | bleb(GO:0032059) |
| 0.2 | 2.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 2.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 2.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 3.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 2.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 2.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 5.4 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.2 | 4.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 3.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.2 | 1.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 8.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.3 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 5.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 12.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 7.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 1.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 4.3 | GO:0016469 | proton-transporting two-sector ATPase complex(GO:0016469) |
| 0.1 | 1.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 1.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 1.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 19.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 10.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 4.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 11.0 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 1.9 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 18.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.9 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 2.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 9.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 8.0 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 2.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 30.0 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 5.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 3.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.3 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 4.0 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 4.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 2.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 1.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 11.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 4.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 1.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 2.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 4.4 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 3.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 9.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 10.3 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 2.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 2.2 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 1.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 3.5 | 10.4 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 3.0 | 14.9 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 3.0 | 8.9 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 2.6 | 7.8 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 1.9 | 5.8 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.8 | 12.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 1.7 | 25.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 1.5 | 19.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 1.5 | 8.8 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 1.4 | 13.9 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 1.4 | 11.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.3 | 35.0 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 1.3 | 3.8 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 1.2 | 4.6 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 1.2 | 24.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 1.1 | 5.7 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 1.1 | 6.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.1 | 3.3 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 1.1 | 4.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.1 | 6.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 1.1 | 7.5 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 1.0 | 8.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.0 | 5.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.0 | 3.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 1.0 | 7.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.9 | 5.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.9 | 17.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.9 | 2.6 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.9 | 2.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.8 | 3.3 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.8 | 4.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.8 | 2.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.8 | 2.3 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.7 | 9.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.7 | 2.1 | GO:0042947 | glucoside transmembrane transporter activity(GO:0042947) |
| 0.7 | 2.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.7 | 4.7 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.7 | 11.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.7 | 2.6 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.6 | 2.6 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.6 | 2.5 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.6 | 3.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.6 | 3.8 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.6 | 2.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.6 | 1.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.6 | 6.6 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.6 | 1.8 | GO:0005135 | erythropoietin receptor binding(GO:0005128) interleukin-3 receptor binding(GO:0005135) |
| 0.6 | 5.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.6 | 1.7 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.6 | 5.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.5 | 9.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.5 | 2.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.5 | 2.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.5 | 4.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.5 | 1.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.5 | 10.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.5 | 7.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.5 | 4.5 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.5 | 3.5 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.5 | 4.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.5 | 17.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.5 | 6.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.5 | 1.4 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.5 | 1.4 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.5 | 11.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.5 | 19.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.5 | 5.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.5 | 3.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.5 | 6.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.5 | 53.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.4 | 4.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 14.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.4 | 6.7 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.4 | 2.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.4 | 6.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 1.3 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.4 | 2.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.4 | 3.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.4 | 3.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.4 | 1.3 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.4 | 3.0 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.4 | 7.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 1.7 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.4 | 8.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 11.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.4 | 1.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 3.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.4 | 5.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 0.8 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.4 | 17.5 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.4 | 2.7 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.4 | 1.1 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.4 | 2.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.4 | 5.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.4 | 5.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.4 | 3.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 6.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.3 | 16.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.3 | 5.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.3 | 1.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.3 | 3.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 11.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.3 | 20.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.3 | 1.3 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.3 | 1.6 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 2.6 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 2.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.3 | 1.9 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 3.7 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.3 | 2.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 16.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 1.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.3 | 1.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.3 | 1.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.3 | 7.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.3 | 6.8 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.3 | 10.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.3 | 2.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 4.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.3 | 2.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.3 | 1.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 1.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.3 | 1.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.3 | 4.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.3 | 1.0 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.3 | 14.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 7.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 5.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 0.8 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 1.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 2.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 3.1 | GO:0090079 | translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.2 | 22.5 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.2 | 0.2 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.2 | 4.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 0.9 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.2 | 8.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 3.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 17.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 4.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 15.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.2 | 0.6 | GO:0052830 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol tetrakisphosphate kinase activity(GO:0051765) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.2 | 8.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 2.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 5.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 12.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 4.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 2.0 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 1.6 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 3.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 2.9 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 1.0 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 4.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 0.6 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.2 | 1.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 3.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 5.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 5.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 1.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 3.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 1.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 1.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 5.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 2.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 1.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 1.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 1.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 0.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 2.0 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.2 | 1.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 1.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 0.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.7 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 2.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 1.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 1.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 6.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 2.7 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.1 | 5.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.9 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 2.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 2.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 13.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 3.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 3.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 1.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.9 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 1.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 2.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 1.0 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 5.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 2.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 3.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.4 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 6.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.8 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 3.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 1.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.7 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 10.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.5 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 3.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 0.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 4.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 8.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 2.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 1.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 2.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 8.1 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 3.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 1.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.3 | GO:0051766 | inositol trisphosphate kinase activity(GO:0051766) |
| 0.1 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.7 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 1.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.6 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 12.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 1.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 3.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 1.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.8 | GO:0071617 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 1.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.7 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.7 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 2.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 1.2 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 1.1 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 3.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.7 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 1.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 25.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.9 | 10.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.8 | 37.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.7 | 14.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 17.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.5 | 13.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.5 | 37.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.5 | 26.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.5 | 35.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.5 | 18.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.4 | 7.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 10.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 10.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 6.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 11.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 7.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 2.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 8.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 19.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 22.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 0.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 9.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.2 | 7.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 9.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 5.8 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.2 | 14.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 3.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 22.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 1.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.2 | 11.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 2.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 5.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 12.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 4.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 13.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 6.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 9.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.9 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 32.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 1.0 | 11.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.9 | 34.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.8 | 10.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.8 | 9.0 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.7 | 30.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.6 | 32.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.6 | 3.4 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.6 | 4.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.5 | 8.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.5 | 4.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.5 | 6.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.5 | 56.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.4 | 1.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.4 | 6.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.4 | 5.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 12.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.4 | 3.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.4 | 6.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 8.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.4 | 12.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.4 | 6.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 13.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.4 | 7.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 7.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.3 | 2.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.3 | 21.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.3 | 3.4 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.3 | 4.5 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.3 | 8.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.3 | 3.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 5.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 5.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 4.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 7.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 18.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 12.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 4.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 1.2 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.2 | 3.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 7.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 4.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 3.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 6.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 7.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 2.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 5.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 4.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 6.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 15.8 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.2 | 7.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 3.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 3.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.2 | 1.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 21.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 4.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 7.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 4.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 8.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 3.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 4.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 24.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 2.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 1.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.9 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.9 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 2.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 2.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 1.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |