Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
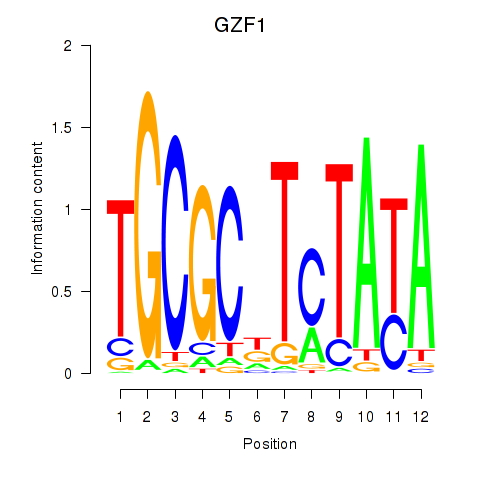
Results for GZF1
Z-value: 3.54
Transcription factors associated with GZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GZF1
|
ENSG00000125812.16 | GZF1 |
Activity profile of GZF1 motif
Sorted Z-values of GZF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GZF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_26280117 | 28.04 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamate rich protein like 3 |
| chr1_+_26280059 | 25.23 |
ENST00000270792.10
|
SH3BGRL3
|
SH3 domain binding glutamate rich protein like 3 |
| chr5_+_171387757 | 22.26 |
ENST00000677297.1
ENST00000521672.6 ENST00000679190.1 ENST00000351986.10 ENST00000676589.1 ENST00000679233.1 ENST00000677357.1 ENST00000296930.10 ENST00000393820.2 ENST00000523622.1 |
NPM1
|
nucleophosmin 1 |
| chr1_+_32292067 | 13.86 |
ENST00000373548.8
ENST00000428704.1 |
HDAC1
|
histone deacetylase 1 |
| chr2_-_149587602 | 13.24 |
ENST00000428879.5
ENST00000303319.10 ENST00000422782.2 |
MMADHC
|
metabolism of cobalamin associated D |
| chr6_-_36547400 | 11.50 |
ENST00000229812.8
|
STK38
|
serine/threonine kinase 38 |
| chr15_+_41286011 | 11.47 |
ENST00000661438.1
|
ENSG00000285920.2
|
novel protein |
| chr7_+_5045821 | 10.65 |
ENST00000353796.7
ENST00000396912.2 ENST00000396904.2 |
RBAK
RBAK-RBAKDN
|
RB associated KRAB zinc finger RBAK-RBAKDN readthrough |
| chr14_+_20989968 | 10.48 |
ENST00000382985.8
ENST00000339374.11 ENST00000556670.6 ENST00000553564.5 ENST00000554751.5 ENST00000554283.1 ENST00000555670.1 |
METTL17
|
methyltransferase like 17 |
| chr12_-_102120065 | 10.44 |
ENST00000552283.6
ENST00000551744.2 |
NUP37
|
nucleoporin 37 |
| chr22_-_42959852 | 8.84 |
ENST00000402229.5
ENST00000407585.5 ENST00000453079.1 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr1_+_78649818 | 8.62 |
ENST00000370747.9
ENST00000438486.1 |
IFI44
|
interferon induced protein 44 |
| chr4_+_26343156 | 8.11 |
ENST00000680928.1
ENST00000681260.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr3_+_30606574 | 7.83 |
ENST00000295754.10
ENST00000359013.4 |
TGFBR2
|
transforming growth factor beta receptor 2 |
| chr6_-_33080710 | 7.31 |
ENST00000419277.5
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr6_+_33080445 | 7.26 |
ENST00000428835.5
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr10_-_102120246 | 6.74 |
ENST00000425280.2
|
LDB1
|
LIM domain binding 1 |
| chr10_+_92593112 | 6.65 |
ENST00000260731.5
|
KIF11
|
kinesin family member 11 |
| chr14_-_21511290 | 6.58 |
ENST00000298717.9
|
METTL3
|
methyltransferase like 3 |
| chr4_-_47463649 | 6.49 |
ENST00000381571.6
|
COMMD8
|
COMM domain containing 8 |
| chr16_+_67842277 | 6.08 |
ENST00000303596.3
|
THAP11
|
THAP domain containing 11 |
| chr7_-_26864573 | 6.05 |
ENST00000345317.7
|
SKAP2
|
src kinase associated phosphoprotein 2 |
| chr1_+_9651723 | 6.00 |
ENST00000377346.9
ENST00000536656.5 ENST00000628140.2 |
PIK3CD
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr10_-_125160499 | 5.77 |
ENST00000494626.6
ENST00000337195.9 |
CTBP2
|
C-terminal binding protein 2 |
| chr10_-_102120318 | 5.53 |
ENST00000673968.1
|
LDB1
|
LIM domain binding 1 |
| chr3_-_79767987 | 5.24 |
ENST00000464233.6
|
ROBO1
|
roundabout guidance receptor 1 |
| chr7_-_93890160 | 5.11 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr11_+_65572349 | 4.95 |
ENST00000316409.2
ENST00000449319.2 ENST00000530349.2 |
FAM89B
|
family with sequence similarity 89 member B |
| chrX_+_81202066 | 4.75 |
ENST00000373212.6
|
SH3BGRL
|
SH3 domain binding glutamate rich protein like |
| chr10_-_125161019 | 4.72 |
ENST00000411419.6
|
CTBP2
|
C-terminal binding protein 2 |
| chr9_+_128371343 | 4.42 |
ENST00000372850.5
ENST00000372847.1 ENST00000372853.9 ENST00000483206.2 |
URM1
|
ubiquitin related modifier 1 |
| chr19_+_926001 | 4.24 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr3_+_51671175 | 4.16 |
ENST00000614067.4
ENST00000457573.5 ENST00000611400.4 ENST00000412249.5 ENST00000425781.5 ENST00000341333.10 ENST00000415259.5 ENST00000395057.5 ENST00000416589.5 |
TEX264
|
testis expressed 264, ER-phagy receptor |
| chr1_-_226186673 | 3.46 |
ENST00000366812.6
|
ACBD3
|
acyl-CoA binding domain containing 3 |
| chr1_+_40709475 | 3.27 |
ENST00000372651.5
|
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr1_+_40709316 | 3.09 |
ENST00000372652.5
|
NFYC
|
nuclear transcription factor Y subunit gamma |
| chrX_+_54809060 | 2.73 |
ENST00000396224.1
|
MAGED2
|
MAGE family member D2 |
| chr10_+_58269132 | 2.38 |
ENST00000333926.6
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr1_-_41918858 | 2.17 |
ENST00000372583.6
|
HIVEP3
|
HIVEP zinc finger 3 |
| chr17_-_31321743 | 2.11 |
ENST00000247270.3
|
EVI2A
|
ecotropic viral integration site 2A |
| chr19_-_3786254 | 2.01 |
ENST00000585778.5
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr19_-_58573555 | 1.69 |
ENST00000599369.5
|
MZF1
|
myeloid zinc finger 1 |
| chr19_-_58573280 | 1.53 |
ENST00000594234.5
ENST00000596039.1 ENST00000215057.7 |
MZF1
|
myeloid zinc finger 1 |
| chr1_-_32336224 | 1.33 |
ENST00000329421.8
|
MARCKSL1
|
MARCKS like 1 |
| chr9_+_134326435 | 1.08 |
ENST00000481739.2
|
RXRA
|
retinoid X receptor alpha |
| chr14_+_49620750 | 1.00 |
ENST00000305386.4
|
MGAT2
|
alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase |
| chr19_+_12995467 | 0.90 |
ENST00000592199.6
|
NFIX
|
nuclear factor I X |
| chr8_-_56113982 | 0.82 |
ENST00000311923.1
|
MOS
|
MOS proto-oncogene, serine/threonine kinase |
| chr19_+_35666515 | 0.74 |
ENST00000617999.4
ENST00000616789.4 |
UPK1A
|
uroplakin 1A |
| chr1_-_41918666 | 0.72 |
ENST00000372584.5
|
HIVEP3
|
HIVEP zinc finger 3 |
| chr19_-_3786408 | 0.40 |
ENST00000395040.6
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr19_+_12995554 | 0.33 |
ENST00000397661.6
|
NFIX
|
nuclear factor I X |
| chr12_+_102120172 | 0.30 |
ENST00000327680.7
ENST00000541394.5 ENST00000543784.5 |
PARPBP
|
PARP1 binding protein |
| chr14_-_52069039 | 0.19 |
ENST00000216286.10
|
NID2
|
nidogen 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.3 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 2.6 | 7.8 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 2.5 | 22.3 | GO:0060699 | regulation of endoribonuclease activity(GO:0060699) |
| 2.0 | 13.9 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 1.7 | 5.0 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 1.3 | 5.2 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 1.2 | 6.0 | GO:0060374 | positive regulation of neutrophil apoptotic process(GO:0033031) mast cell differentiation(GO:0060374) |
| 1.2 | 8.1 | GO:1901189 | arterial endothelial cell fate commitment(GO:0060844) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 1.1 | 4.4 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 1.1 | 6.6 | GO:1903679 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 1.0 | 10.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.9 | 8.8 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.7 | 53.3 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.5 | 13.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.4 | 2.7 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.4 | 1.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.3 | 6.7 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.3 | 0.8 | GO:0051295 | meiotic cell cycle phase transition(GO:0044771) establishment of meiotic spindle localization(GO:0051295) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.2 | 10.4 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 14.6 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.1 | 5.1 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 10.0 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.1 | 6.4 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 2.4 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 1.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 4.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 2.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 10.5 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 4.2 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 3.5 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 6.1 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 1.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 6.4 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 14.4 | GO:0045892 | negative regulation of transcription, DNA-templated(GO:0045892) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.6 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 1.3 | 7.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.1 | 22.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.9 | 8.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.8 | 14.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.7 | 13.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.7 | 10.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.6 | 6.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.6 | 10.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 6.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 50.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 8.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 6.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 6.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 6.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 12.3 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 10.5 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 4.2 | GO:0045121 | membrane raft(GO:0045121) |
| 0.0 | 2.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 22.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 3.3 | 53.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 2.6 | 7.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 2.2 | 6.6 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 1.5 | 10.5 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 1.4 | 12.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.9 | 13.9 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.7 | 5.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.6 | 11.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.6 | 6.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.5 | 7.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.5 | 8.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.4 | 1.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.3 | 8.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.3 | 4.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.3 | 5.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 6.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 7.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 3.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 2.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 10.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 6.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 9.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 5.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 10.5 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 10.6 | GO:0008270 | zinc ion binding(GO:0008270) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 13.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.4 | 22.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 6.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 7.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 8.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 8.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 5.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 10.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 22.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.7 | 14.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 8.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.4 | 13.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.4 | 7.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.3 | 10.4 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.2 | 6.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 5.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 6.0 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.2 | 6.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 6.6 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.0 | 1.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.0 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |