Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for HES1
Z-value: 7.85
Transcription factors associated with HES1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HES1
|
ENSG00000114315.4 | HES1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HES1 | hg38_v1_chr3_+_194136138_194136155 | -0.18 | 8.3e-03 | Click! |
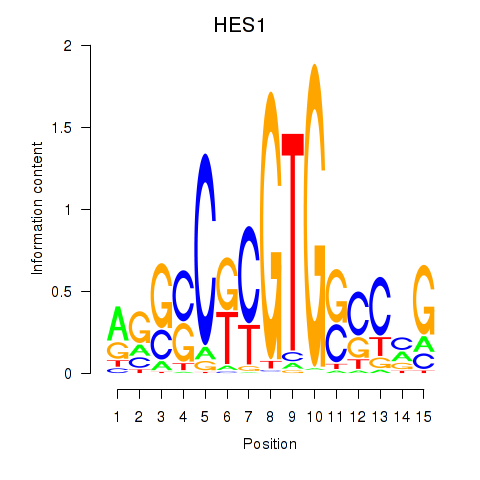
Activity profile of HES1 motif
Sorted Z-values of HES1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HES1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 42.7 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 8.2 | 41.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 6.3 | 18.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 5.2 | 15.7 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 5.2 | 15.6 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 5.1 | 15.3 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 4.8 | 38.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 4.0 | 15.9 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 3.9 | 15.7 | GO:2000426 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 3.6 | 10.9 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 3.6 | 21.6 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 3.3 | 3.3 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 3.0 | 12.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 3.0 | 17.9 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 2.9 | 8.8 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 2.9 | 11.5 | GO:0009216 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) |
| 2.8 | 8.5 | GO:0097086 | amniotic stem cell differentiation(GO:0097086) negative regulation of dense core granule biogenesis(GO:2000706) negative regulation of mesenchymal stem cell differentiation(GO:2000740) regulation of amniotic stem cell differentiation(GO:2000797) negative regulation of amniotic stem cell differentiation(GO:2000798) |
| 2.7 | 8.2 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) mast cell proliferation(GO:0070662) |
| 2.7 | 8.0 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 2.7 | 18.7 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 2.6 | 10.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 2.5 | 7.5 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 2.4 | 7.3 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 2.4 | 41.0 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 2.4 | 7.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 2.3 | 7.0 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 2.3 | 11.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 2.2 | 8.8 | GO:0002188 | translation reinitiation(GO:0002188) |
| 2.2 | 19.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 2.1 | 6.4 | GO:0097069 | response to human chorionic gonadotropin(GO:0044752) cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 2.1 | 8.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 2.1 | 6.3 | GO:0006208 | purine nucleobase catabolic process(GO:0006145) pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) |
| 2.1 | 16.5 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 2.0 | 18.0 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 2.0 | 5.9 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 1.9 | 5.8 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 1.9 | 9.6 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 1.9 | 5.6 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) |
| 1.8 | 7.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 1.8 | 5.3 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 1.8 | 10.6 | GO:0098838 | methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 1.8 | 36.9 | GO:0006206 | pyrimidine nucleobase metabolic process(GO:0006206) |
| 1.7 | 20.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.7 | 69.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 1.7 | 8.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 1.7 | 5.0 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 1.6 | 13.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 1.6 | 7.9 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 1.6 | 6.3 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 1.6 | 6.3 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 1.6 | 4.7 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 1.5 | 10.8 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 1.5 | 4.5 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 1.5 | 7.4 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 1.4 | 8.6 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 1.4 | 5.8 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 1.4 | 4.2 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 1.4 | 18.1 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 1.4 | 4.1 | GO:1902594 | viral penetration into host nucleus(GO:0075732) multi-organism nuclear import(GO:1902594) |
| 1.4 | 4.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 1.3 | 17.0 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 1.3 | 3.9 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 1.3 | 10.3 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 1.2 | 9.9 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 1.2 | 6.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 1.2 | 21.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 1.2 | 4.7 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 1.1 | 4.6 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 1.1 | 6.8 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 1.1 | 15.9 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 1.1 | 25.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 1.1 | 13.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 1.1 | 5.5 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 1.1 | 5.5 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 1.1 | 3.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 1.1 | 5.5 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 1.1 | 14.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 1.1 | 13.7 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 1.0 | 7.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 1.0 | 4.0 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 1.0 | 14.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 1.0 | 7.0 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 1.0 | 3.0 | GO:1902595 | DNA replication preinitiation complex assembly(GO:0071163) regulation of DNA replication origin binding(GO:1902595) |
| 1.0 | 5.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 1.0 | 8.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 1.0 | 5.9 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 1.0 | 10.5 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 1.0 | 8.6 | GO:0090151 | protein import into mitochondrial inner membrane(GO:0045039) establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.9 | 4.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.9 | 10.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.9 | 3.7 | GO:0003290 | septum secundum development(GO:0003285) atrial septum secundum morphogenesis(GO:0003290) |
| 0.9 | 5.4 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.9 | 1.8 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.9 | 26.0 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.9 | 2.7 | GO:0036233 | glycine import(GO:0036233) |
| 0.9 | 2.7 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.9 | 3.5 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.9 | 3.5 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.9 | 9.5 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.9 | 6.8 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.8 | 3.4 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.8 | 5.8 | GO:0032328 | alanine transport(GO:0032328) |
| 0.8 | 24.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.8 | 4.0 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.8 | 23.0 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.8 | 3.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.8 | 4.6 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.8 | 3.1 | GO:0021634 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.8 | 18.8 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.8 | 15.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.7 | 1.5 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.7 | 3.0 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.7 | 10.8 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.7 | 5.8 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.7 | 19.3 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.7 | 6.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.7 | 6.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.7 | 1.3 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.6 | 3.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.6 | 3.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.6 | 10.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.6 | 3.7 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.6 | 3.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.6 | 5.0 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.6 | 1.8 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.6 | 7.3 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.6 | 3.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.6 | 1.2 | GO:0071287 | cellular response to manganese ion(GO:0071287) |
| 0.6 | 2.9 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.6 | 4.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.6 | 2.9 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.6 | 21.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.6 | 3.9 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.6 | 4.4 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.5 | 13.2 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.5 | 3.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.5 | 2.7 | GO:1902268 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.5 | 12.8 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.5 | 2.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.5 | 8.7 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.5 | 12.9 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.5 | 4.2 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.5 | 2.3 | GO:0097021 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.5 | 1.4 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.5 | 2.3 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.4 | 2.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.4 | 1.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.4 | 8.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.4 | 8.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 5.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.4 | 2.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.4 | 3.8 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 15.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.4 | 7.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.4 | 2.0 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.4 | 2.4 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.4 | 3.5 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.4 | 1.2 | GO:0060957 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 0.4 | 13.0 | GO:0032094 | response to food(GO:0032094) |
| 0.4 | 3.3 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.4 | 6.9 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.4 | 3.9 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.3 | 4.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.3 | 3.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.3 | 3.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.3 | 3.0 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 1.3 | GO:0071110 | protein biotinylation(GO:0009305) histone biotinylation(GO:0071110) |
| 0.3 | 1.9 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.3 | 33.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.3 | 2.8 | GO:0019043 | establishment of viral latency(GO:0019043) establishment of integrated proviral latency(GO:0075713) |
| 0.3 | 1.9 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.3 | 2.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.3 | 0.9 | GO:0072366 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.3 | 4.7 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.3 | 3.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.3 | 9.0 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.3 | 1.1 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.3 | 4.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.3 | 7.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.3 | 5.5 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.3 | 6.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.2 | 1.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.2 | 0.7 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.2 | 2.2 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.2 | 5.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) mitotic G2/M transition checkpoint(GO:0044818) |
| 0.2 | 4.1 | GO:0098969 | neurotransmitter receptor transport to plasma membrane(GO:0098877) neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.2 | 8.7 | GO:0006101 | citrate metabolic process(GO:0006101) |
| 0.2 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 1.7 | GO:0001878 | response to yeast(GO:0001878) |
| 0.2 | 2.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 1.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 7.7 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.2 | 1.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 0.9 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.2 | 12.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 11.6 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.2 | 1.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 1.5 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 2.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 3.5 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 7.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.2 | 1.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 4.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.2 | 2.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 1.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 | 1.0 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 2.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.2 | 1.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 | 5.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.2 | 11.0 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.2 | 1.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 5.4 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.2 | 7.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.2 | 17.2 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.2 | 9.8 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.2 | 6.6 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.2 | 13.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 1.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 3.1 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.2 | 2.3 | GO:0009112 | nucleobase metabolic process(GO:0009112) |
| 0.1 | 23.8 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 0.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.4 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.4 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 7.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 1.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 3.5 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 0.5 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.4 | GO:0090176 | microtubule cytoskeleton organization involved in establishment of planar polarity(GO:0090176) |
| 0.1 | 0.6 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 3.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 7.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 3.6 | GO:0072663 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 2.2 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.1 | 8.1 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 3.9 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 4.0 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 8.1 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 12.3 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.1 | 1.2 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 4.7 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.1 | 8.8 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 1.3 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.1 | 5.7 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 7.7 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 9.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 3.9 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 1.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 8.6 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.1 | 1.5 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.1 | 2.5 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.1 | 1.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 12.1 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.1 | 5.5 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 1.8 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 4.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 22.1 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.1 | 0.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 7.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 2.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 2.7 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 0.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 1.3 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.1 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 1.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 1.9 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.1 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 2.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 3.6 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.1 | 3.0 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.1 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 1.3 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 0.2 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 1.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:1904782 | negative regulation of glutamate receptor signaling pathway(GO:1900450) negative regulation of NMDA glutamate receptor activity(GO:1904782) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.4 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.7 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 1.9 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 1.8 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 6.8 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 1.3 | GO:0051149 | positive regulation of muscle cell differentiation(GO:0051149) |
| 0.0 | 3.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 1.2 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.4 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.1 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 14.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 4.0 | 64.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 3.8 | 11.3 | GO:0044393 | microspike(GO:0044393) |
| 3.3 | 36.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 3.0 | 8.9 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 2.5 | 12.4 | GO:0089701 | U2AF(GO:0089701) |
| 2.4 | 26.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 2.3 | 9.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 2.2 | 15.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 2.2 | 12.9 | GO:0001940 | male pronucleus(GO:0001940) |
| 2.1 | 6.4 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 1.9 | 9.7 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 1.8 | 14.0 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 1.7 | 20.3 | GO:0042555 | MCM complex(GO:0042555) |
| 1.6 | 8.2 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 1.6 | 9.7 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 1.5 | 5.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.5 | 7.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) myosin II filament(GO:0097513) |
| 1.4 | 8.7 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 1.4 | 7.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 1.3 | 38.7 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 1.3 | 11.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 1.2 | 34.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 1.2 | 25.6 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 1.2 | 15.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 1.2 | 10.7 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 1.2 | 3.5 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 1.2 | 5.8 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 1.1 | 3.3 | GO:0000805 | X chromosome(GO:0000805) |
| 1.1 | 9.9 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 1.1 | 10.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 1.1 | 6.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 1.0 | 5.0 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 1.0 | 11.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 1.0 | 5.8 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 1.0 | 3.8 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.9 | 6.6 | GO:0032021 | NELF complex(GO:0032021) |
| 0.9 | 51.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.8 | 4.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.8 | 7.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.8 | 6.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.8 | 16.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.8 | 6.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.7 | 3.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.7 | 16.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.7 | 1.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.7 | 12.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.7 | 7.3 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.7 | 6.6 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.7 | 3.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.6 | 5.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.6 | 32.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.6 | 9.1 | GO:0001741 | XY body(GO:0001741) |
| 0.6 | 5.9 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.6 | 3.5 | GO:0005816 | spindle pole body(GO:0005816) |
| 0.6 | 4.0 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.6 | 6.8 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.6 | 27.6 | GO:0005844 | polysome(GO:0005844) |
| 0.5 | 4.4 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.5 | 3.7 | GO:1990745 | EARP complex(GO:1990745) |
| 0.5 | 2.5 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.5 | 6.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.5 | 4.9 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.5 | 6.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.5 | 12.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.5 | 2.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.5 | 13.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.5 | 8.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.5 | 3.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.5 | 11.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.5 | 2.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.4 | 3.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 5.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.4 | 84.1 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.4 | 1.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.4 | 10.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.4 | 1.2 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.3 | 6.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 10.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.3 | 1.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.3 | 8.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.3 | 14.1 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.3 | 3.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.3 | 19.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 3.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 3.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.3 | 14.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.3 | 27.5 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.3 | 22.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 2.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.3 | 23.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 3.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 13.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 7.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 4.2 | GO:0031932 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.2 | 1.5 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 16.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 4.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 3.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 15.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 3.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 34.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.2 | 7.3 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.2 | 30.4 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 8.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 7.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 51.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 2.9 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.9 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 43.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 7.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 2.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 5.4 | GO:0032153 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.1 | 9.1 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 1.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 3.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 7.3 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 1.2 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.4 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.1 | 0.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 11.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 1.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 3.7 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 2.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 2.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.3 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 18.1 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.8 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 2.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 41.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 6.3 | 31.7 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 6.3 | 18.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 6.1 | 42.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 5.2 | 15.7 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 4.6 | 9.3 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 4.0 | 64.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 3.4 | 36.9 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 3.2 | 29.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 3.0 | 12.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 3.0 | 8.9 | GO:0004638 | phosphoribosylaminoimidazole carboxylase activity(GO:0004638) phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 2.9 | 8.8 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 2.9 | 11.5 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 2.8 | 17.0 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 2.7 | 35.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 2.6 | 28.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 2.3 | 14.0 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 2.2 | 8.7 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 2.2 | 8.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 2.1 | 6.4 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 2.1 | 6.4 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 2.1 | 8.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 2.0 | 5.9 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 1.9 | 21.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 1.9 | 5.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 1.9 | 24.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 1.8 | 10.6 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 1.7 | 6.9 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 1.7 | 6.8 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 1.7 | 8.3 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 1.6 | 8.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 1.6 | 11.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 1.6 | 6.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.6 | 4.7 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 1.5 | 7.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 1.5 | 11.7 | GO:0050733 | RS domain binding(GO:0050733) |
| 1.4 | 15.9 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 1.4 | 5.8 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 1.4 | 12.9 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 1.4 | 4.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 1.4 | 6.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 1.3 | 8.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 1.3 | 3.9 | GO:0016662 | oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 1.2 | 7.5 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 1.2 | 19.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 1.2 | 7.4 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 1.2 | 31.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 1.2 | 3.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 1.1 | 3.4 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 1.1 | 5.6 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 1.1 | 8.7 | GO:0015288 | porin activity(GO:0015288) |
| 1.0 | 7.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 1.0 | 3.1 | GO:0000248 | C-5 sterol desaturase activity(GO:0000248) sterol desaturase activity(GO:0070704) |
| 1.0 | 5.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 1.0 | 5.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 1.0 | 15.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 1.0 | 14.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 1.0 | 8.9 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 1.0 | 9.9 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 1.0 | 6.9 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.0 | 5.8 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 1.0 | 3.9 | GO:0004146 | dihydrofolate reductase activity(GO:0004146) |
| 1.0 | 4.8 | GO:0031762 | alpha-1A adrenergic receptor binding(GO:0031691) follicle-stimulating hormone receptor binding(GO:0031762) |
| 1.0 | 3.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.9 | 4.7 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.9 | 2.7 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.8 | 11.0 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.8 | 4.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.8 | 3.3 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.8 | 9.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.8 | 1.6 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.8 | 3.9 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.8 | 35.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.7 | 5.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.7 | 4.9 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.7 | 47.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.7 | 2.8 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.7 | 4.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.7 | 2.6 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.7 | 5.9 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.7 | 34.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.6 | 6.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.6 | 6.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.6 | 5.6 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.6 | 8.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.6 | 30.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.6 | 14.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.5 | 3.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.5 | 3.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.5 | 2.7 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.5 | 1.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.5 | 6.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.5 | 3.7 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.5 | 12.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.5 | 1.5 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.5 | 3.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.5 | 2.9 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.5 | 11.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.5 | 13.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.5 | 30.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.5 | 1.9 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.5 | 5.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.5 | 2.3 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.4 | 11.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 17.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.4 | 13.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.4 | 6.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.4 | 18.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.4 | 3.7 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.4 | 2.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.4 | 3.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 2.9 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.4 | 1.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.4 | 3.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.4 | 10.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 6.9 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.4 | 10.2 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.4 | 7.4 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.4 | 5.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 11.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.3 | 6.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.3 | 6.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 1.3 | GO:0004078 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.3 | 5.5 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.3 | 10.8 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.3 | 3.7 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.3 | 13.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.3 | 8.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 3.9 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.3 | 16.0 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.3 | 4.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 9.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.3 | 4.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.3 | 7.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.3 | 7.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.3 | 0.8 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 1.1 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) |
| 0.3 | 2.7 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.3 | 14.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.3 | 5.0 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.2 | 3.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 1.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 16.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.2 | 3.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 4.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 3.6 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.2 | 3.1 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.2 | 5.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 6.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.2 | 8.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 11.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.2 | 2.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 15.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.2 | 2.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 3.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 1.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 1.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 1.2 | GO:0034617 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) tetrahydrobiopterin binding(GO:0034617) |
| 0.2 | 0.7 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.2 | 3.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 35.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.2 | 3.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 27.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 10.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 1.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 5.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 5.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 3.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.3 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 1.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 1.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 5.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 1.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 6.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 5.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 4.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 3.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 3.4 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.1 | 0.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 2.2 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 2.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 2.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.7 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 3.9 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.1 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 4.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 0.2 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.1 | 4.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 6.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 5.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.8 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 12.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 2.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 4.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 1.2 | GO:0016859 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.0 | 1.0 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.9 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 4.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 36.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.7 | 29.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.7 | 95.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.4 | 9.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.4 | 14.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.4 | 17.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.4 | 7.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.4 | 10.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.4 | 29.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.4 | 15.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.4 | 16.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 13.1 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 14.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.3 | 13.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.3 | 13.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.3 | 11.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.3 | 18.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 11.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 13.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 14.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 1.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 2.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 1.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 13.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 11.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 12.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 2.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 7.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 7.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 6.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 6.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 8.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 3.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 2.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 37.7 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 3.2 | 63.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 3.1 | 64.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 3.0 | 3.0 | REACTOME M G1 TRANSITION | Genes involved in M/G1 Transition |
| 2.3 | 20.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 1.6 | 23.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 1.6 | 23.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.5 | 58.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 1.2 | 17.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 1.1 | 17.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 1.1 | 11.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 1.0 | 9.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.0 | 3.9 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.9 | 6.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.8 | 6.8 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.7 | 12.5 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.7 | 9.8 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.6 | 13.8 | REACTOME PURINE METABOLISM | Genes involved in Purine metabolism |
| 0.6 | 15.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.6 | 10.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.6 | 7.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.6 | 26.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.6 | 9.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.6 | 10.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.5 | 27.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.5 | 8.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.5 | 15.9 | REACTOME METABOLISM OF NUCLEOTIDES | Genes involved in Metabolism of nucleotides |
| 0.5 | 30.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.5 | 10.8 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.5 | 24.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.5 | 47.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.5 | 2.8 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.5 | 6.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.4 | 3.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.4 | 5.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.4 | 7.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 92.3 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.4 | 49.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.3 | 2.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.3 | 8.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 5.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 6.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.3 | 19.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 23.5 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.3 | 15.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.2 | 21.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 5.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 6.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 8.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 10.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 3.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.2 | 5.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 8.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 13.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 10.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 3.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 4.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 2.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 28.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 5.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 1.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 7.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 12.9 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 4.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 5.0 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.1 | 1.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 2.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 2.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.2 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 2.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 3.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 4.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |