Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
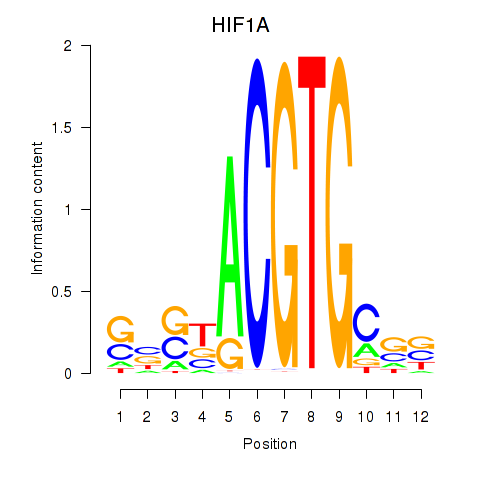
Results for HIF1A
Z-value: 4.68
Transcription factors associated with HIF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIF1A
|
ENSG00000100644.17 | HIF1A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIF1A | hg38_v1_chr14_+_61695435_61695598, hg38_v1_chr14_+_61695777_61695813, hg38_v1_chr14_+_61697622_61697717 | 0.62 | 1.5e-24 | Click! |
Activity profile of HIF1A motif
Sorted Z-values of HIF1A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HIF1A
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.1 | 100.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 5.8 | 17.5 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 4.5 | 18.0 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 4.2 | 12.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 3.8 | 15.3 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 3.8 | 45.5 | GO:1905098 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 3.7 | 14.9 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 3.4 | 16.9 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 2.9 | 8.8 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 2.8 | 17.0 | GO:0002424 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 2.8 | 16.7 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 2.7 | 8.2 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 2.7 | 10.9 | GO:0006175 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) |
| 2.5 | 12.7 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 2.5 | 19.8 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 2.5 | 7.4 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 2.4 | 9.7 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 2.4 | 7.1 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 2.4 | 7.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.3 | 7.0 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 2.2 | 6.7 | GO:0097698 | telomere maintenance via base-excision repair(GO:0097698) |
| 2.1 | 6.4 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 2.1 | 10.3 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 2.0 | 17.8 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 1.9 | 7.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.9 | 5.6 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 1.8 | 16.5 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 1.8 | 22.8 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 1.7 | 5.2 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 1.7 | 7.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 1.7 | 5.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 1.6 | 7.8 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 1.5 | 16.9 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 1.5 | 10.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) negative regulation of megakaryocyte differentiation(GO:0045653) |
| 1.4 | 7.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 1.4 | 7.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 1.3 | 8.1 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 1.3 | 22.5 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 1.3 | 3.9 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 1.3 | 23.1 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 1.3 | 3.8 | GO:0021592 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 1.2 | 3.7 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 1.2 | 7.0 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 1.2 | 16.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 1.1 | 8.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 1.1 | 12.0 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 1.1 | 6.4 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 1.0 | 20.1 | GO:0031639 | plasminogen activation(GO:0031639) |
| 1.0 | 6.9 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.9 | 3.5 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.9 | 2.6 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.8 | 6.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.8 | 2.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.7 | 11.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.7 | 2.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.6 | 11.0 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.6 | 3.8 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.6 | 10.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.6 | 4.3 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.6 | 2.3 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.6 | 2.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.6 | 2.8 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.5 | 1.6 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.5 | 3.6 | GO:0015677 | copper ion import(GO:0015677) |
| 0.5 | 9.8 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.5 | 14.5 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.5 | 3.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.5 | 3.9 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.5 | 1.4 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.5 | 4.8 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.5 | 7.6 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.5 | 3.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 4.8 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.4 | 1.3 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.4 | 1.7 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.4 | 7.1 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.4 | 8.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.4 | 7.9 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.4 | 3.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.4 | 1.5 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.4 | 11.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) error-free translesion synthesis(GO:0070987) |
| 0.3 | 31.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.3 | 6.9 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.3 | 4.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 8.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.3 | 2.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 3.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 1.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.3 | 3.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 3.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 1.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.2 | 16.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 13.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 21.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.2 | 1.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 0.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 | 0.6 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.2 | 9.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.2 | 12.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.2 | 0.4 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 1.7 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 0.8 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 7.8 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.2 | 7.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.2 | 12.5 | GO:0035904 | aorta development(GO:0035904) |
| 0.2 | 10.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.2 | 2.7 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 4.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 1.7 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 2.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 2.7 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 2.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 6.8 | GO:0018208 | protein peptidyl-prolyl isomerization(GO:0000413) peptidyl-proline modification(GO:0018208) |
| 0.1 | 0.4 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.3 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 1.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 6.4 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.1 | 0.4 | GO:1904799 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 | 6.4 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.1 | 0.7 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 1.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 5.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.6 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 6.2 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 1.6 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.1 | 2.6 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.9 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.3 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 1.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 7.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 9.0 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 3.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.2 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 1.7 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.2 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 1.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 2.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 10.1 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 2.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.6 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 11.3 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 2.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 3.7 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 1.4 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 3.2 | GO:2001020 | regulation of response to DNA damage stimulus(GO:2001020) |
| 0.0 | 19.8 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.5 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.5 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0043418 | cysteine biosynthetic process from serine(GO:0006535) cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) homocysteine catabolic process(GO:0043418) L-cysteine metabolic process(GO:0046439) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.5 | 100.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 3.6 | 14.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 3.2 | 12.7 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 2.8 | 16.9 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 2.1 | 16.9 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 1.7 | 20.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.6 | 19.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 1.6 | 17.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 1.5 | 16.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 1.4 | 19.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 1.3 | 10.3 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 1.2 | 8.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 1.1 | 6.8 | GO:0070545 | PeBoW complex(GO:0070545) |
| 1.1 | 9.7 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.0 | 5.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 1.0 | 26.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 1.0 | 9.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.9 | 16.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.9 | 10.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.9 | 12.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.8 | 5.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.8 | 7.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) lipopolysaccharide receptor complex(GO:0046696) |
| 0.8 | 3.8 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.7 | 17.0 | GO:0046930 | pore complex(GO:0046930) |
| 0.7 | 18.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.6 | 10.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.6 | 14.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.6 | 10.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.5 | 4.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.5 | 8.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.5 | 7.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.5 | 2.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.4 | 16.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.4 | 6.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 1.0 | GO:0044393 | microspike(GO:0044393) |
| 0.3 | 21.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.3 | 31.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 2.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 3.9 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.2 | 1.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 9.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 9.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 15.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 11.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 6.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 2.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 42.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 7.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 2.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 4.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 4.3 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 2.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 11.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 5.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 2.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 5.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.4 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 3.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 6.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 30.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 12.0 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 3.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 2.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 11.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 6.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 81.5 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 3.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 2.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 2.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.8 | GO:0005840 | ribosome(GO:0005840) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.5 | 100.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 7.7 | 23.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 6.7 | 20.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 4.5 | 18.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 4.2 | 12.7 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 4.2 | 12.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 4.2 | 16.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 3.2 | 9.7 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 3.2 | 22.5 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 2.7 | 10.9 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 2.5 | 10.1 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) |
| 2.5 | 19.8 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 2.4 | 12.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 2.4 | 7.1 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 2.4 | 7.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 2.1 | 16.9 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 2.1 | 16.9 | GO:0015288 | porin activity(GO:0015288) |
| 2.0 | 21.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 1.7 | 7.0 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 1.7 | 10.3 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 1.7 | 6.7 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 1.6 | 7.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 1.6 | 4.7 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.5 | 8.8 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 1.4 | 7.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.4 | 9.9 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 1.4 | 14.9 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 1.3 | 7.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 1.2 | 12.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 1.2 | 4.8 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 1.2 | 7.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 1.1 | 10.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 1.0 | 8.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.9 | 28.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.9 | 3.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.7 | 3.7 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.7 | 12.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.7 | 12.5 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.7 | 19.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.7 | 3.3 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.6 | 8.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.6 | 8.8 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.6 | 1.7 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.6 | 19.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.6 | 1.7 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.6 | 27.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.6 | 14.5 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.5 | 1.5 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.5 | 2.9 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.5 | 13.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.5 | 2.3 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.5 | 6.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 7.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.4 | 7.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.4 | 1.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.4 | 16.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.4 | 15.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.4 | 11.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.4 | 2.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 10.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 5.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.4 | 9.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.4 | 4.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.4 | 2.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.4 | 6.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.4 | 26.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.4 | 2.5 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 2.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 2.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 1.3 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.3 | 3.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.3 | 7.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 2.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 6.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 2.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 6.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 46.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 6.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 13.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 0.8 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 4.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 7.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.2 | 0.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 2.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.2 | 3.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 2.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.4 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 7.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 6.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 2.7 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 1.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 9.2 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 0.5 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 13.5 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 1.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 8.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 2.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 2.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 32.0 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 1.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 3.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.7 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 8.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 7.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 0.2 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 4.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 2.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 8.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 5.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.8 | GO:0016896 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0050421 | oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.0 | 1.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 49.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.3 | 182.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.8 | 42.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.5 | 44.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.5 | 12.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 9.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 16.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 2.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 8.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 21.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 3.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 5.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 25.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 5.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 6.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 6.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 3.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 8.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.0 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 146.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 1.3 | 38.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.8 | 28.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.7 | 8.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.7 | 25.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.7 | 9.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.6 | 19.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.6 | 18.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.6 | 8.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.6 | 13.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.6 | 18.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 37.5 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.5 | 14.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.4 | 9.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.4 | 31.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.4 | 11.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 23.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 9.8 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.3 | 7.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 9.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 31.0 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.3 | 9.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.3 | 18.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.3 | 18.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 8.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 9.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 2.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 24.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 3.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 6.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 4.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 6.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 3.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 12.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 15.1 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.1 | 3.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 10.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 5.3 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 2.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.2 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 3.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 5.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 3.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 7.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.4 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |