Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
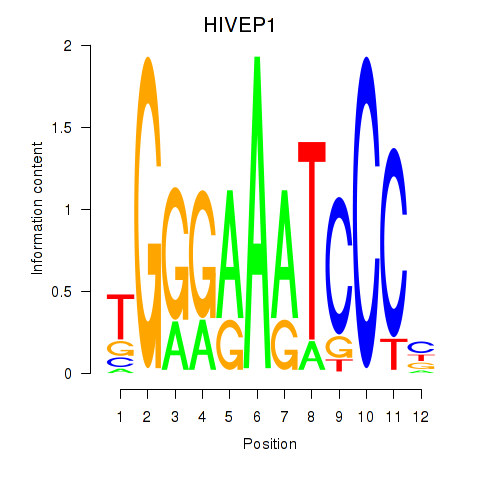
Results for HIVEP1
Z-value: 5.44
Transcription factors associated with HIVEP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIVEP1
|
ENSG00000095951.17 | HIVEP1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIVEP1 | hg38_v1_chr6_+_12012304_12012338 | 0.14 | 4.0e-02 | Click! |
Activity profile of HIVEP1 motif
Sorted Z-values of HIVEP1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HIVEP1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.6 | 67.7 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 17.7 | 53.2 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 11.5 | 34.4 | GO:0034147 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 8.9 | 62.6 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 8.5 | 84.5 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 6.1 | 24.5 | GO:0045401 | response to molecule of fungal origin(GO:0002238) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) cellular response to molecule of fungal origin(GO:0071226) |
| 5.8 | 17.5 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 5.5 | 16.4 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 5.4 | 65.4 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 5.4 | 16.3 | GO:0036292 | DNA rewinding(GO:0036292) |
| 5.1 | 15.3 | GO:1900226 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 4.5 | 18.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 3.7 | 15.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 3.7 | 33.6 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 3.4 | 17.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 3.4 | 10.2 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 3.0 | 9.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 2.6 | 20.8 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 2.6 | 28.5 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 2.4 | 7.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 2.4 | 19.2 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 2.3 | 9.3 | GO:0043335 | protein unfolding(GO:0043335) |
| 2.2 | 6.7 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 2.2 | 6.6 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 2.1 | 8.4 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 2.1 | 6.2 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 2.1 | 154.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 2.0 | 28.5 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 2.0 | 6.0 | GO:0060018 | basophil differentiation(GO:0030221) positive regulation of chromatin assembly or disassembly(GO:0045799) astrocyte fate commitment(GO:0060018) |
| 1.9 | 7.8 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 1.9 | 24.4 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 1.7 | 1.7 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 1.5 | 4.5 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 1.4 | 7.2 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 1.4 | 4.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 1.4 | 9.5 | GO:0036018 | response to cobalamin(GO:0033590) cellular response to erythropoietin(GO:0036018) |
| 1.3 | 5.4 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 1.3 | 2.6 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 1.2 | 3.7 | GO:0045425 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 1.2 | 12.8 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.2 | 4.6 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 1.1 | 9.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.1 | 9.8 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 1.1 | 3.2 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 1.1 | 3.2 | GO:1904747 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 1.1 | 3.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 1.1 | 21.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 1.1 | 21.1 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 1.0 | 102.0 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 1.0 | 4.1 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 1.0 | 3.0 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 1.0 | 116.3 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.9 | 2.7 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.9 | 9.5 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.8 | 2.5 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.8 | 3.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.8 | 7.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.8 | 16.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.8 | 10.0 | GO:0032329 | serine transport(GO:0032329) |
| 0.8 | 5.4 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.8 | 1.5 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.7 | 7.4 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.7 | 2.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.7 | 5.7 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.7 | 4.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.7 | 20.0 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.7 | 68.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.7 | 2.1 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) regulation of chromatin silencing at telomere(GO:0031938) |
| 0.7 | 10.9 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.7 | 6.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.7 | 4.6 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.7 | 3.3 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) positive regulation of cellular pH reduction(GO:0032849) |
| 0.6 | 7.7 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.6 | 7.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.6 | 11.4 | GO:0042832 | defense response to protozoan(GO:0042832) T-helper 2 cell differentiation(GO:0045064) |
| 0.6 | 5.6 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.6 | 3.7 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.6 | 5.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.6 | 6.5 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.6 | 8.0 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.6 | 2.8 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.6 | 2.8 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.6 | 6.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.5 | 8.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.5 | 12.9 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.5 | 2.5 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.5 | 3.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.5 | 9.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.5 | 7.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.5 | 6.0 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.5 | 5.4 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.4 | 3.0 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.4 | 3.0 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.4 | 2.1 | GO:0046641 | positive regulation of alpha-beta T cell proliferation(GO:0046641) |
| 0.4 | 2.5 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.4 | 5.5 | GO:0048535 | lymph node development(GO:0048535) |
| 0.4 | 2.1 | GO:1902430 | negative regulation of beta-amyloid formation(GO:1902430) |
| 0.4 | 2.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.4 | 1.3 | GO:0052314 | serotonin biosynthetic process(GO:0042427) phytoalexin metabolic process(GO:0052314) |
| 0.4 | 3.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.4 | 1.2 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.4 | 8.1 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.4 | 3.7 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.4 | 1.5 | GO:2000909 | negative regulation of triglyceride catabolic process(GO:0010897) regulation of high-density lipoprotein particle clearance(GO:0010982) negative regulation of low-density lipoprotein particle clearance(GO:0010989) regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.4 | 5.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 6.9 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.3 | 3.4 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.3 | 1.5 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.3 | 12.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.3 | 3.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 2.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 2.9 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.3 | 7.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 1.5 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.2 | 18.3 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.2 | 1.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.2 | 0.7 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.2 | 1.1 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.2 | 4.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 5.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 4.7 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.2 | 1.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 1.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 1.5 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.2 | 1.5 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.2 | 2.5 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.2 | 16.9 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.2 | 1.6 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.2 | 0.8 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.2 | 4.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 1.6 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.2 | 6.9 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.2 | 6.2 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 1.7 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 4.5 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 24.2 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.2 | 0.9 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.2 | 4.0 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.1 | 4.0 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 2.8 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 1.3 | GO:1901898 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 4.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 3.6 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 4.1 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.1 | 4.9 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.1 | 4.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 5.8 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 4.6 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.1 | 4.7 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 6.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.6 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.1 | 1.9 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 1.7 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 1.8 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 7.2 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.1 | 4.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.5 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 1.7 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.1 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 3.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 2.1 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.1 | 3.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 2.9 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.1 | 1.9 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 3.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 6.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 2.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.7 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 0.2 | GO:0046645 | positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.1 | 1.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 3.9 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.1 | 2.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.8 | GO:0097118 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.1 | GO:2000077 | negative regulation of dopaminergic neuron differentiation(GO:1904339) negative regulation of type B pancreatic cell development(GO:2000077) negative regulation of glycogen (starch) synthase activity(GO:2000466) |
| 0.1 | 1.2 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 4.1 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 1.3 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.1 | 3.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 4.5 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 3.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.0 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.1 | 6.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.3 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 4.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 11.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 0.6 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 2.1 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.1 | 1.1 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 5.9 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 0.8 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 12.7 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.1 | 5.0 | GO:0006024 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 3.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 4.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 2.2 | GO:0032642 | regulation of chemokine production(GO:0032642) |
| 0.0 | 0.4 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 1.3 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 3.2 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.0 | 0.6 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 2.1 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 3.8 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.5 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 8.0 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 2.3 | GO:0007004 | telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.9 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 2.3 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.0 | 0.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.2 | GO:0034311 | sphingosine metabolic process(GO:0006670) diol metabolic process(GO:0034311) |
| 0.0 | 1.0 | GO:1902400 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 1.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 1.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.8 | GO:0098773 | skin epidermis development(GO:0098773) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.6 | 220.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 7.7 | 84.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 7.7 | 23.0 | GO:0071754 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 4.9 | 24.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 3.3 | 43.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 2.6 | 131.4 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 2.3 | 20.3 | GO:0032010 | phagolysosome(GO:0032010) |
| 2.2 | 6.7 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 2.1 | 46.0 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 2.1 | 16.4 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 1.9 | 7.8 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 1.7 | 15.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 1.6 | 6.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.6 | 4.9 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 1.6 | 17.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.5 | 6.0 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 1.2 | 17.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.1 | 8.0 | GO:0043196 | varicosity(GO:0043196) |
| 1.1 | 12.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 1.1 | 18.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 1.0 | 18.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 1.0 | 9.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.9 | 6.5 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.9 | 8.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.9 | 4.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.8 | 21.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.8 | 5.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.7 | 2.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.7 | 12.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.6 | 8.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.6 | 4.0 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.6 | 3.4 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.6 | 6.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.5 | 3.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.5 | 10.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.5 | 4.7 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.5 | 6.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.5 | 9.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.5 | 5.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.5 | 4.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.5 | 4.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.5 | 5.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.4 | 4.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.4 | 8.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.4 | 2.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.4 | 27.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.4 | 5.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.3 | 5.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 16.3 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.3 | 2.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 1.9 | GO:0060091 | kinocilium(GO:0060091) |
| 0.3 | 25.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.3 | 3.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.3 | 1.8 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 11.1 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 8.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 23.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 0.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 3.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 1.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.2 | 1.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 2.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 1.5 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 3.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 25.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.2 | 0.9 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 1.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 15.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 2.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 2.8 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 4.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 8.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 6.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 10.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 41.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.5 | GO:0035841 | growing cell tip(GO:0035838) new growing cell tip(GO:0035841) cell tip(GO:0051286) |
| 0.1 | 4.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 3.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 2.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 3.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 3.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 16.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 83.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 35.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 15.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 4.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 2.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 5.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 6.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 2.4 | GO:0034358 | plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.1 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 10.3 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 5.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 2.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 16.6 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 2.4 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 1.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 149.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 10.6 | 53.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 6.4 | 38.6 | GO:0046979 | TAP2 binding(GO:0046979) |
| 5.1 | 15.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 4.9 | 24.4 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 4.1 | 16.3 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 2.6 | 154.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 2.5 | 20.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 2.5 | 34.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 2.1 | 19.2 | GO:0043426 | MRF binding(GO:0043426) |
| 1.9 | 5.7 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 1.8 | 45.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 1.7 | 17.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 1.5 | 9.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 1.4 | 24.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 1.4 | 9.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.3 | 18.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 1.3 | 9.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.3 | 18.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 1.3 | 19.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.2 | 4.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 1.2 | 3.7 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 1.2 | 12.8 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 1.1 | 8.0 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 1.1 | 3.4 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 1.1 | 4.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.1 | 20.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 1.1 | 3.2 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.9 | 6.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.9 | 2.8 | GO:0042806 | fucose binding(GO:0042806) |
| 0.9 | 2.8 | GO:0016154 | thymidine phosphorylase activity(GO:0009032) pyrimidine-nucleoside phosphorylase activity(GO:0016154) |
| 0.9 | 9.9 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.9 | 5.1 | GO:0016416 | O-palmitoyltransferase activity(GO:0016416) |
| 0.8 | 4.9 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.8 | 14.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.8 | 3.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.7 | 3.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.7 | 5.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.7 | 25.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.7 | 23.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.7 | 9.5 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.7 | 2.9 | GO:0019862 | IgA binding(GO:0019862) |
| 0.7 | 3.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.7 | 5.4 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.6 | 3.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.6 | 10.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.6 | 88.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.6 | 1.8 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.6 | 6.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.6 | 1.7 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.6 | 6.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.5 | 8.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.5 | 6.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.5 | 2.9 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.5 | 6.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.4 | 24.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.4 | 2.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.4 | 7.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.4 | 3.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 7.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.4 | 4.7 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.4 | 12.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.4 | 6.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.4 | 3.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.4 | 1.8 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.4 | 9.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.4 | 12.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.4 | 4.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 7.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.3 | 5.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.3 | 26.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.3 | 1.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.3 | 11.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 2.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 1.3 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.3 | 22.0 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.3 | 5.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 1.5 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.3 | 4.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.3 | 1.7 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.3 | 6.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 3.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 4.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.3 | 5.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 1.5 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.3 | 4.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 6.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 4.2 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.2 | 1.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 7.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 4.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 7.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 1.3 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 0.8 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 4.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.2 | 2.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 7.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.2 | 1.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 2.8 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 2.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 5.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 1.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 0.5 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.2 | 2.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 3.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 3.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 32.0 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.1 | 5.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 4.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 7.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 2.1 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 3.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.0 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 23.4 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.1 | 9.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 3.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 7.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 4.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 2.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 3.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 3.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 21.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 1.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 6.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 3.1 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 1.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 9.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 5.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.0 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 1.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 4.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 1.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.2 | GO:0042166 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 8.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 1.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 3.5 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 2.2 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 3.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 2.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 6.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.7 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 62.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 2.0 | 64.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 1.3 | 64.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 1.1 | 53.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 1.0 | 23.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 1.0 | 53.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.8 | 34.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.7 | 15.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.7 | 30.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.6 | 13.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.5 | 17.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.4 | 5.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.4 | 18.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.3 | 3.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.3 | 11.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 6.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 11.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 6.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 3.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 6.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 9.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.2 | 3.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 1.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 4.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.2 | 4.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 4.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 7.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 7.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 17.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 7.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 6.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 6.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 2.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 5.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 4.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 16.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 4.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 84.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 3.8 | 79.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 3.1 | 64.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 2.4 | 17.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 1.1 | 15.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 1.0 | 20.6 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.9 | 44.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.8 | 70.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.8 | 35.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.7 | 9.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.7 | 17.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.6 | 12.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.5 | 12.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.5 | 68.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.4 | 6.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 9.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 13.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.4 | 11.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.4 | 13.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 4.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 5.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 6.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.3 | 5.6 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.3 | 4.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 6.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 5.7 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.3 | 4.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 2.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 13.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 10.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 3.8 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.2 | 5.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 4.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 3.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 2.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 4.3 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.2 | 6.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.2 | 11.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 5.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.7 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 4.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 3.1 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 6.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 4.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 9.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 6.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 2.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 4.8 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 1.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.2 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 3.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 3.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 6.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 3.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 4.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 10.5 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |