Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
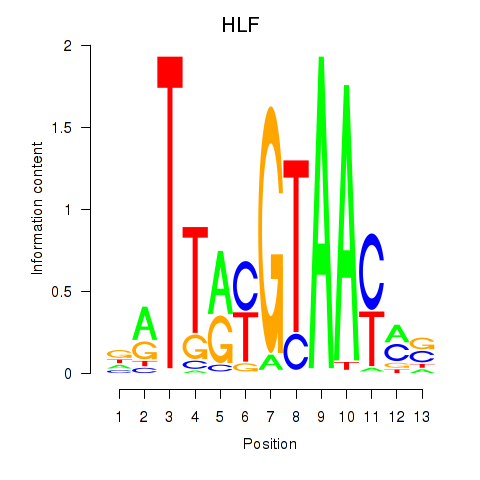
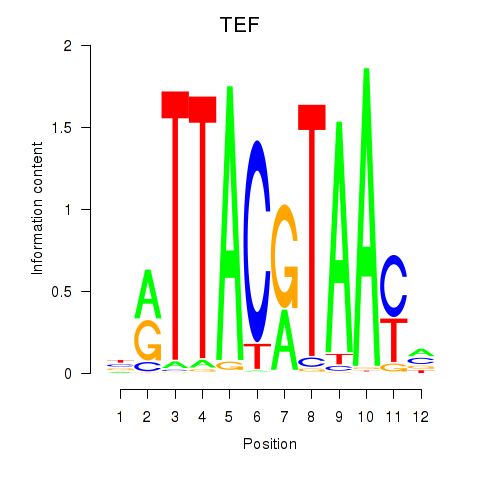
Results for HLF_TEF
Z-value: 7.14
Transcription factors associated with HLF_TEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HLF
|
ENSG00000108924.14 | HLF |
|
TEF
|
ENSG00000167074.15 | TEF |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEF | hg38_v1_chr22_+_41381923_41381968, hg38_v1_chr22_+_41367269_41367333 | -0.66 | 2.5e-28 | Click! |
| HLF | hg38_v1_chr17_+_55267584_55267616 | -0.50 | 3.4e-15 | Click! |
Activity profile of HLF_TEF motif
Sorted Z-values of HLF_TEF motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HLF_TEF
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 150.9 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 6.8 | 27.0 | GO:1990535 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 5.7 | 17.1 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 5.5 | 16.4 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 4.6 | 27.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 2.5 | 12.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 2.5 | 34.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 2.4 | 19.4 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 2.1 | 6.4 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 2.1 | 16.8 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 2.0 | 46.6 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 2.0 | 6.1 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 1.9 | 22.3 | GO:0006983 | ER overload response(GO:0006983) |
| 1.8 | 8.9 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.7 | 7.0 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 1.6 | 6.4 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 1.1 | 10.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.1 | 4.5 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 1.1 | 4.3 | GO:1903786 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 1.0 | 14.7 | GO:0070487 | monocyte aggregation(GO:0070487) |
| 1.0 | 5.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 1.0 | 7.0 | GO:0015866 | ADP transport(GO:0015866) |
| 1.0 | 7.9 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 1.0 | 7.9 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.9 | 88.5 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.9 | 9.2 | GO:0045078 | enucleate erythrocyte differentiation(GO:0043353) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.9 | 8.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.8 | 14.3 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.8 | 5.6 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.7 | 3.0 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.7 | 2.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.7 | 5.0 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.6 | 1.9 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.6 | 5.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.5 | 7.8 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.5 | 3.6 | GO:0090204 | protein localization to nuclear pore(GO:0090204) positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.4 | 9.7 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.4 | 2.5 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.4 | 4.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.4 | 3.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.4 | 6.6 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.4 | 24.5 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.4 | 30.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.4 | 1.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 0.3 | 3.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.3 | 1.9 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.3 | 2.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.3 | 12.1 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.2 | 7.3 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.2 | 0.8 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.2 | 1.5 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.2 | 8.0 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.2 | 4.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.2 | 12.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.2 | 3.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 4.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 1.2 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 0.6 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.1 | 4.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 2.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.5 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 1.7 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 2.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 1.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 3.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 10.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 12.8 | GO:0051495 | positive regulation of cytoskeleton organization(GO:0051495) |
| 0.0 | 0.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.8 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 7.5 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 1.0 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 1.0 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.2 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 1.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 9.0 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 1.6 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 1.3 | GO:0070268 | cornification(GO:0070268) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 34.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 3.3 | 13.3 | GO:0071942 | XPC complex(GO:0071942) |
| 2.7 | 27.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 2.1 | 16.4 | GO:0005827 | polar microtubule(GO:0005827) |
| 2.0 | 14.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 1.3 | 7.9 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 1.2 | 12.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.9 | 21.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.9 | 3.6 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.8 | 30.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.7 | 91.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.6 | 1.9 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.6 | 9.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.5 | 8.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.4 | 7.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.4 | 8.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.4 | 5.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 3.0 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.3 | 34.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 11.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 5.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 7.3 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.2 | 12.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 1.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 1.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 21.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 6.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 6.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.0 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 11.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 9.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 11.1 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 6.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 3.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 2.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 31.5 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 2.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 19.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 11.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 4.3 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 4.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 8.8 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 6.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 11.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 8.1 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 103.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 5.7 | 17.1 | GO:0031403 | lithium ion binding(GO:0031403) |
| 5.5 | 16.4 | GO:0031208 | POZ domain binding(GO:0031208) |
| 5.4 | 27.0 | GO:1903135 | cupric ion binding(GO:1903135) |
| 4.8 | 14.3 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 4.7 | 46.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 4.6 | 27.3 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 2.8 | 16.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 2.3 | 7.0 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 1.3 | 10.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 1.1 | 6.6 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 1.0 | 5.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 1.0 | 7.0 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 1.0 | 3.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.9 | 34.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.8 | 4.2 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.8 | 4.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.8 | 8.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.7 | 23.0 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.6 | 3.0 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.5 | 7.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.5 | 6.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.5 | 9.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.5 | 1.4 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.4 | 7.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.4 | 5.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.4 | 14.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.4 | 13.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 12.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 1.9 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.3 | 2.5 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.3 | 5.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 2.8 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.3 | 6.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.3 | 6.1 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.3 | 9.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 1.6 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 5.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 1.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 4.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 2.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 13.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 3.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 7.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 4.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 30.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 3.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 4.3 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 0.6 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 1.7 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 38.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 9.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 1.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 7.3 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 4.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 4.9 | GO:0015405 | primary active transmembrane transporter activity(GO:0015399) P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 24.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 4.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 4.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 27.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.4 | 16.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.4 | 14.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 23.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 13.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 3.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 10.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 5.8 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 4.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 7.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 5.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 5.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 4.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 8.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 8.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 3.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 88.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.9 | 19.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.7 | 8.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.6 | 16.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.5 | 12.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.5 | 11.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.4 | 13.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.4 | 6.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.4 | 10.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 14.1 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.3 | 23.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 7.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 5.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.3 | 17.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 8.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 7.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 3.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 5.8 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 2.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 9.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 11.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 9.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 12.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |