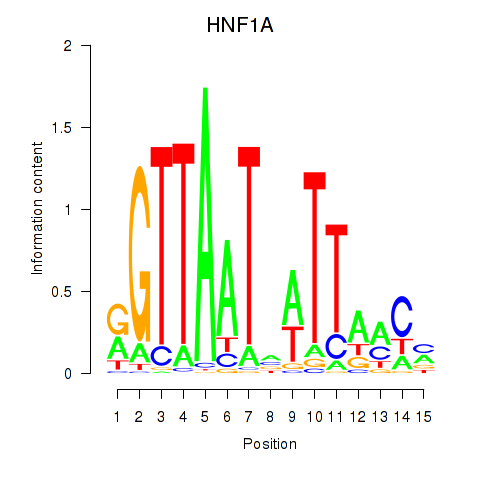
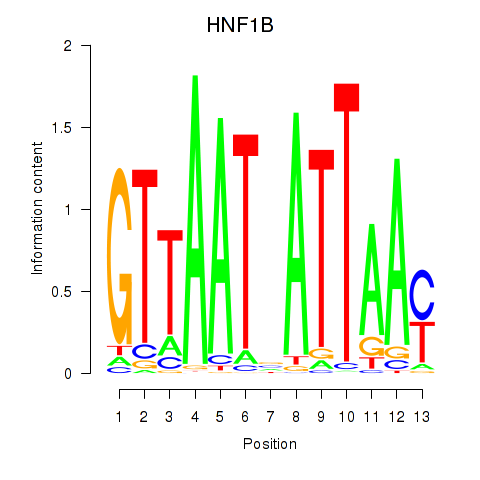
|
chr17_-_66229380
Show fit
|
61.70 |
ENST00000205948.11
|
APOH
|
apolipoprotein H
|
|
chr4_-_71784046
Show fit
|
60.20 |
ENST00000513476.5
ENST00000273951.13
|
GC
|
GC vitamin D binding protein
|
|
chr4_+_73436198
Show fit
|
50.59 |
ENST00000395792.7
|
AFP
|
alpha fetoprotein
|
|
chr4_+_73436244
Show fit
|
50.18 |
ENST00000226359.2
|
AFP
|
alpha fetoprotein
|
|
chr6_+_160702238
Show fit
|
41.49 |
ENST00000366924.6
ENST00000308192.14
ENST00000418964.1
|
PLG
|
plasminogen
|
|
chr4_+_154563003
Show fit
|
39.31 |
ENST00000302068.9
ENST00000509493.1
|
FGB
|
fibrinogen beta chain
|
|
chr19_-_35812838
Show fit
|
36.87 |
ENST00000653904.2
|
PRODH2
|
proline dehydrogenase 2
|
|
chr14_-_94388589
Show fit
|
35.71 |
ENST00000402629.1
ENST00000556091.1
ENST00000393087.9
ENST00000554720.1
|
SERPINA1
|
serpin family A member 1
|
|
chr4_-_154590735
Show fit
|
34.86 |
ENST00000403106.8
ENST00000622532.1
ENST00000651975.1
|
FGA
|
fibrinogen alpha chain
|
|
chr4_+_73404255
Show fit
|
33.24 |
ENST00000621628.4
ENST00000621085.4
ENST00000415165.6
ENST00000295897.9
ENST00000503124.5
ENST00000509063.5
ENST00000401494.7
|
ALB
|
albumin
|
|
chr2_+_233760265
Show fit
|
32.96 |
ENST00000305208.10
ENST00000360418.4
|
UGT1A1
|
UDP glucuronosyltransferase family 1 member A1
|
|
chr2_+_233692881
Show fit
|
32.06 |
ENST00000305139.11
|
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6
|
|
chr4_+_73481737
Show fit
|
31.88 |
ENST00000226355.5
|
AFM
|
afamin
|
|
chr9_-_121050264
Show fit
|
28.47 |
ENST00000223642.3
|
C5
|
complement C5
|
|
chr15_+_58431985
Show fit
|
28.13 |
ENST00000433326.2
ENST00000299022.10
|
LIPC
|
lipase C, hepatic type
|
|
chr7_-_50565381
Show fit
|
27.92 |
ENST00000444124.7
|
DDC
|
dopa decarboxylase
|
|
chr16_-_20576277
Show fit
|
27.41 |
ENST00000566384.5
ENST00000565232.5
ENST00000567001.5
ENST00000565322.5
ENST00000569344.5
ENST00000329697.10
ENST00000568882.1
|
ACSM2B
|
acyl-CoA synthetase medium chain family member 2B
|
|
chr1_+_207104226
Show fit
|
22.40 |
ENST00000367070.8
|
C4BPA
|
complement component 4 binding protein alpha
|
|
chr19_+_49513353
Show fit
|
22.03 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter
|
|
chr6_-_160664270
Show fit
|
20.74 |
ENST00000316300.10
|
LPA
|
lipoprotein(a)
|
|
chr7_+_45888360
Show fit
|
20.70 |
ENST00000457280.5
|
IGFBP1
|
insulin like growth factor binding protein 1
|
|
chr16_+_20451461
Show fit
|
20.55 |
ENST00000574251.5
ENST00000573854.6
|
ACSM2A
|
acyl-CoA synthetase medium chain family member 2A
|
|
chr8_-_17895487
Show fit
|
20.46 |
ENST00000427924.5
ENST00000381841.4
|
FGL1
|
fibrinogen like 1
|
|
chr2_-_88128049
Show fit
|
20.17 |
ENST00000393750.3
ENST00000295834.8
|
FABP1
|
fatty acid binding protein 1
|
|
chr16_+_20451596
Show fit
|
19.93 |
ENST00000575690.5
ENST00000571894.1
|
ACSM2A
|
acyl-CoA synthetase medium chain family member 2A
|
|
chr16_+_20451563
Show fit
|
19.55 |
ENST00000417235.6
ENST00000219054.10
|
ACSM2A
|
acyl-CoA synthetase medium chain family member 2A
|
|
chr14_-_94293071
Show fit
|
19.47 |
ENST00000554723.5
|
SERPINA10
|
serpin family A member 10
|
|
chr14_-_94293024
Show fit
|
19.44 |
ENST00000393096.5
|
SERPINA10
|
serpin family A member 10
|
|
chr2_+_233729042
Show fit
|
19.38 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3
|
|
chr19_+_49513154
Show fit
|
18.48 |
ENST00000426395.7
ENST00000600273.5
ENST00000599988.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter
|
|
chr7_+_45888479
Show fit
|
18.24 |
ENST00000275525.8
ENST00000468955.1
|
IGFBP1
|
insulin like growth factor binding protein 1
|
|
chr10_+_113553039
Show fit
|
17.88 |
ENST00000351270.4
|
HABP2
|
hyaluronan binding protein 2
|
|
chr5_+_177086867
Show fit
|
17.71 |
ENST00000503708.5
ENST00000393648.6
ENST00000514472.1
ENST00000502906.5
ENST00000292408.9
ENST00000510911.5
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr14_-_94293260
Show fit
|
17.70 |
ENST00000261994.9
|
SERPINA10
|
serpin family A member 10
|
|
chr6_+_31655888
Show fit
|
17.55 |
ENST00000375916.4
|
APOM
|
apolipoprotein M
|
|
chr2_+_87748087
Show fit
|
17.20 |
ENST00000359481.9
|
PLGLB2
|
plasminogen like B2
|
|
chr3_+_186666003
Show fit
|
16.73 |
ENST00000232003.5
|
HRG
|
histidine rich glycoprotein
|
|
chr4_+_3441960
Show fit
|
16.06 |
ENST00000382774.8
ENST00000511533.1
|
HGFAC
|
HGF activator
|
|
chr14_+_94561435
Show fit
|
15.69 |
ENST00000557004.6
ENST00000555095.5
ENST00000298841.5
ENST00000554220.5
ENST00000553780.5
|
SERPINA4
SERPINA5
|
serpin family A member 4
serpin family A member 5
|
|
chr14_-_57493827
Show fit
|
15.47 |
ENST00000526336.1
ENST00000216445.8
|
CCDC198
|
coiled-coil domain containing 198
|
|
chr6_-_25930611
Show fit
|
15.03 |
ENST00000360488.7
|
SLC17A2
|
solute carrier family 17 member 2
|
|
chr19_+_11239602
Show fit
|
14.95 |
ENST00000252453.12
|
ANGPTL8
|
angiopoietin like 8
|
|
chr14_+_20688756
Show fit
|
14.83 |
ENST00000397990.5
ENST00000555597.1
|
ANG
RNASE4
|
angiogenin
ribonuclease A family member 4
|
|
chr17_+_42900791
Show fit
|
14.70 |
ENST00000592383.5
ENST00000253801.7
ENST00000585489.1
|
G6PC1
|
glucose-6-phosphatase catalytic subunit 1
|
|
chr6_-_25930678
Show fit
|
14.24 |
ENST00000377850.8
|
SLC17A2
|
solute carrier family 17 member 2
|
|
chr11_+_64555684
Show fit
|
14.08 |
ENST00000377585.7
|
SLC22A11
|
solute carrier family 22 member 11
|
|
chr17_+_1742836
Show fit
|
13.85 |
ENST00000324015.7
ENST00000450523.6
ENST00000453723.5
ENST00000453066.6
ENST00000382061.5
|
SERPINF2
|
serpin family F member 2
|
|
chr20_+_43558968
Show fit
|
13.31 |
ENST00000647834.1
ENST00000373100.7
ENST00000648083.1
ENST00000648530.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr2_-_87021844
Show fit
|
13.26 |
ENST00000355705.4
ENST00000409310.6
|
PLGLB1
|
plasminogen like B1
|
|
chr10_+_133527355
Show fit
|
13.20 |
ENST00000252945.8
ENST00000421586.5
ENST00000418356.1
|
CYP2E1
|
cytochrome P450 family 2 subfamily E member 1
|
|
chr11_-_117824734
Show fit
|
13.10 |
ENST00000292079.7
|
FXYD2
|
FXYD domain containing ion transport regulator 2
|
|
chr12_-_91180365
Show fit
|
13.10 |
ENST00000547937.5
|
DCN
|
decorin
|
|
chr7_-_44541318
Show fit
|
13.07 |
ENST00000381160.8
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1
|
|
chr2_+_233617626
Show fit
|
12.83 |
ENST00000373450.5
|
UGT1A8
|
UDP glucuronosyltransferase family 1 member A8
|
|
chr12_-_91153149
Show fit
|
12.60 |
ENST00000550758.1
|
DCN
|
decorin
|
|
chr4_-_99290975
Show fit
|
12.57 |
ENST00000209668.3
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
|
|
chr11_+_46719193
Show fit
|
12.05 |
ENST00000311907.10
ENST00000530231.5
ENST00000442468.1
|
F2
|
coagulation factor II, thrombin
|
|
chr11_+_114296347
Show fit
|
11.78 |
ENST00000299964.4
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr11_+_64555924
Show fit
|
11.55 |
ENST00000301891.9
|
SLC22A11
|
solute carrier family 22 member 11
|
|
chr6_-_52763473
Show fit
|
11.54 |
ENST00000493422.3
|
GSTA2
|
glutathione S-transferase alpha 2
|
|
chr1_+_17580474
Show fit
|
11.51 |
ENST00000375415.5
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor 10 like
|
|
chrX_-_106038721
Show fit
|
11.49 |
ENST00000372563.2
|
SERPINA7
|
serpin family A member 7
|
|
chr7_-_44541262
Show fit
|
11.30 |
ENST00000289547.8
ENST00000546276.5
ENST00000423141.1
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1
|
|
chr1_+_103750406
Show fit
|
11.26 |
ENST00000370079.3
|
AMY1C
|
amylase alpha 1C
|
|
chr4_-_99352754
Show fit
|
10.68 |
ENST00000639454.1
|
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr2_+_233718734
Show fit
|
10.49 |
ENST00000373409.8
|
UGT1A4
|
UDP glucuronosyltransferase family 1 member A4
|
|
chr20_+_44355692
Show fit
|
10.12 |
ENST00000316673.8
ENST00000609795.5
ENST00000457232.5
ENST00000609262.5
|
HNF4A
|
hepatocyte nuclear factor 4 alpha
|
|
chr16_+_20764036
Show fit
|
10.07 |
ENST00000440284.6
|
ACSM3
|
acyl-CoA synthetase medium chain family member 3
|
|
chr16_-_66918839
Show fit
|
10.01 |
ENST00000565235.2
ENST00000568632.5
ENST00000565796.5
|
CDH16
|
cadherin 16
|
|
chr22_-_30574572
Show fit
|
10.01 |
ENST00000402369.5
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1
|
|
chr16_-_66918876
Show fit
|
9.94 |
ENST00000570262.5
ENST00000299752.9
ENST00000394055.7
|
CDH16
|
cadherin 16
|
|
chr8_-_69833338
Show fit
|
9.92 |
ENST00000524945.5
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1
|
|
chr21_-_46155567
Show fit
|
9.89 |
ENST00000291670.9
ENST00000397748.5
ENST00000397743.1
ENST00000397746.8
|
FTCD
|
formimidoyltransferase cyclodeaminase
|
|
chr10_+_99782628
Show fit
|
8.56 |
ENST00000648689.1
ENST00000647814.1
|
ABCC2
|
ATP binding cassette subfamily C member 2
|
|
chr16_+_20763990
Show fit
|
8.33 |
ENST00000289416.10
|
ACSM3
|
acyl-CoA synthetase medium chain family member 3
|
|
chr20_-_7940444
Show fit
|
8.25 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1
|
|
chr4_+_99574812
Show fit
|
8.09 |
ENST00000422897.6
ENST00000265517.10
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr10_-_50885619
Show fit
|
8.04 |
ENST00000373997.8
|
A1CF
|
APOBEC1 complementation factor
|
|
chr10_-_50885656
Show fit
|
7.91 |
ENST00000374001.6
ENST00000395489.6
ENST00000282641.6
ENST00000395495.5
ENST00000373995.7
ENST00000414883.1
|
A1CF
|
APOBEC1 complementation factor
|
|
chr2_+_233712905
Show fit
|
7.78 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5
|
|
chr6_+_25754699
Show fit
|
7.55 |
ENST00000439485.6
ENST00000377905.9
|
SLC17A4
|
solute carrier family 17 member 4
|
|
chr17_-_352784
Show fit
|
7.47 |
ENST00000577079.5
ENST00000331302.12
ENST00000618002.4
ENST00000536489.6
|
RPH3AL
|
rabphilin 3A like (without C2 domains)
|
|
chr2_-_227379297
Show fit
|
7.44 |
ENST00000304568.4
|
TM4SF20
|
transmembrane 4 L six family member 20
|
|
chr8_-_94208548
Show fit
|
7.33 |
ENST00000027335.8
ENST00000441892.6
ENST00000521491.1
|
CDH17
|
cadherin 17
|
|
chr1_+_119368773
Show fit
|
7.17 |
ENST00000457318.5
ENST00000622548.4
ENST00000325945.4
|
HAO2
|
hydroxyacid oxidase 2
|
|
chr6_+_131250375
Show fit
|
6.95 |
ENST00000474850.2
|
AKAP7
|
A-kinase anchoring protein 7
|
|
chr4_-_68670648
Show fit
|
6.92 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15
|
|
chr4_-_99352730
Show fit
|
6.75 |
ENST00000510055.5
ENST00000515683.6
ENST00000511397.3
|
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide
|
|
chr5_+_176549323
Show fit
|
6.70 |
ENST00000261944.9
|
CDHR2
|
cadherin related family member 2
|
|
chr17_+_28473278
Show fit
|
6.65 |
ENST00000444914.7
|
SLC13A2
|
solute carrier family 13 member 2
|
|
chr7_+_141995872
Show fit
|
6.65 |
ENST00000497673.5
ENST00000620571.1
ENST00000475668.6
|
MGAM
|
maltase-glucoamylase
|
|
chr4_-_1173168
Show fit
|
6.61 |
ENST00000514490.5
ENST00000431380.5
ENST00000503765.5
|
SPON2
|
spondin 2
|
|
chr12_-_102480638
Show fit
|
6.38 |
ENST00000392904.5
|
IGF1
|
insulin like growth factor 1
|
|
chr1_-_19815270
Show fit
|
6.22 |
ENST00000375121.4
|
RNF186
|
ring finger protein 186
|
|
chr1_+_62597510
Show fit
|
6.09 |
ENST00000371129.4
|
ANGPTL3
|
angiopoietin like 3
|
|
chr4_-_109801978
Show fit
|
5.92 |
ENST00000510800.1
ENST00000512148.5
ENST00000394634.7
ENST00000394635.8
ENST00000645635.1
|
CFI
ENSG00000285330.1
|
complement factor I
novel protein
|
|
chr21_+_38256698
Show fit
|
5.90 |
ENST00000613499.4
ENST00000612702.4
ENST00000398925.5
ENST00000398928.5
ENST00000328656.8
ENST00000443341.5
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15
|
|
chr6_-_25874212
Show fit
|
5.86 |
ENST00000361703.10
ENST00000397060.8
|
SLC17A3
|
solute carrier family 17 member 3
|
|
chr7_+_141995826
Show fit
|
5.81 |
ENST00000549489.6
|
MGAM
|
maltase-glucoamylase
|
|
chr15_+_48206286
Show fit
|
5.78 |
ENST00000396577.7
ENST00000380993.8
|
SLC12A1
|
solute carrier family 12 member 1
|
|
chr9_-_91950201
Show fit
|
5.72 |
ENST00000375708.4
|
ROR2
|
receptor tyrosine kinase like orphan receptor 2
|
|
chr1_+_56854764
Show fit
|
5.71 |
ENST00000361249.4
|
C8A
|
complement C8 alpha chain
|
|
chr20_+_44401397
Show fit
|
5.68 |
ENST00000682427.1
ENST00000681977.1
ENST00000684136.1
ENST00000684046.1
ENST00000684476.1
ENST00000619550.5
ENST00000682169.1
|
HNF4A
|
hepatocyte nuclear factor 4 alpha
|
|
chrX_-_15601077
Show fit
|
5.67 |
ENST00000680121.1
|
ACE2
|
angiotensin I converting enzyme 2
|
|
chr13_+_33016415
Show fit
|
5.45 |
ENST00000380099.4
|
KL
|
klotho
|
|
chr12_-_102480604
Show fit
|
5.32 |
ENST00000392905.7
|
IGF1
|
insulin like growth factor 1
|
|
chr17_+_28473635
Show fit
|
5.30 |
ENST00000314669.10
ENST00000545060.2
|
SLC13A2
|
solute carrier family 13 member 2
|
|
chr2_-_162074394
Show fit
|
5.29 |
ENST00000676810.1
|
DPP4
|
dipeptidyl peptidase 4
|
|
chr12_+_100473916
Show fit
|
5.28 |
ENST00000549996.5
|
NR1H4
|
nuclear receptor subfamily 1 group H member 4
|
|
chr12_-_102480552
Show fit
|
5.26 |
ENST00000337514.11
ENST00000307046.8
|
IGF1
|
insulin like growth factor 1
|
|
chr12_+_100503352
Show fit
|
5.19 |
ENST00000551379.5
ENST00000188403.7
|
NR1H4
|
nuclear receptor subfamily 1 group H member 4
|
|
chr19_+_1205761
Show fit
|
5.15 |
ENST00000326873.12
ENST00000586243.5
|
STK11
|
serine/threonine kinase 11
|
|
chr6_+_167291329
Show fit
|
5.00 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A
|
|
chr19_+_41797147
Show fit
|
4.99 |
ENST00000596544.1
|
CEACAM3
|
CEA cell adhesion molecule 3
|
|
chr21_+_38256984
Show fit
|
4.85 |
ENST00000398938.7
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15
|
|
chr14_+_64704380
Show fit
|
4.84 |
ENST00000247226.13
ENST00000394691.7
|
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3
|
|
chr2_-_162074182
Show fit
|
4.71 |
ENST00000360534.8
|
DPP4
|
dipeptidyl peptidase 4
|
|
chr16_+_89630263
Show fit
|
4.66 |
ENST00000261615.5
|
DPEP1
|
dipeptidase 1
|
|
chrX_-_15600953
Show fit
|
4.66 |
ENST00000679212.1
ENST00000679278.1
ENST00000678046.1
ENST00000252519.8
|
ACE2
|
angiotensin I converting enzyme 2
|
|
chr2_-_162074050
Show fit
|
4.62 |
ENST00000676768.1
|
DPP4
|
dipeptidyl peptidase 4
|
|
chr8_-_86743626
Show fit
|
4.56 |
ENST00000320005.6
|
CNGB3
|
cyclic nucleotide gated channel subunit beta 3
|
|
chr7_-_27180230
Show fit
|
4.55 |
ENST00000396344.4
|
HOXA10
|
homeobox A10
|
|
chr12_+_100473951
Show fit
|
4.35 |
ENST00000648861.1
ENST00000546380.1
|
NR1H4
|
nuclear receptor subfamily 1 group H member 4
|
|
chr14_-_64942720
Show fit
|
4.27 |
ENST00000557049.1
ENST00000389614.6
|
GPX2
|
glutathione peroxidase 2
|
|
chr3_-_138174879
Show fit
|
4.24 |
ENST00000260803.9
|
DBR1
|
debranching RNA lariats 1
|
|
chr6_-_116060859
Show fit
|
4.17 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase
|
|
chr22_+_44702186
Show fit
|
4.14 |
ENST00000336985.11
ENST00000403696.5
ENST00000457960.5
ENST00000361473.9
|
PRR5
PRR5-ARHGAP8
|
proline rich 5
PRR5-ARHGAP8 readthrough
|
|
chr4_+_186266183
Show fit
|
4.11 |
ENST00000403665.7
ENST00000492972.6
ENST00000264692.8
|
F11
|
coagulation factor XI
|
|
chr2_+_27496830
Show fit
|
4.09 |
ENST00000264717.7
|
GCKR
|
glucokinase regulator
|
|
chr6_+_31739948
Show fit
|
4.05 |
ENST00000375755.8
ENST00000425703.5
ENST00000375750.9
ENST00000375703.7
ENST00000375740.7
|
MSH5
|
mutS homolog 5
|
|
chr12_+_21131187
Show fit
|
3.96 |
ENST00000256958.3
|
SLCO1B1
|
solute carrier organic anion transporter family member 1B1
|
|
chr14_-_64942783
Show fit
|
3.92 |
ENST00000612794.1
|
GPX2
|
glutathione peroxidase 2
|
|
chr14_+_39233908
Show fit
|
3.91 |
ENST00000280082.4
|
MIA2
|
MIA SH3 domain ER export factor 2
|
|
chr8_+_22053543
Show fit
|
3.86 |
ENST00000519850.5
ENST00000381470.7
|
DMTN
|
dematin actin binding protein
|
|
chrX_+_16650155
Show fit
|
3.84 |
ENST00000380200.3
|
S100G
|
S100 calcium binding protein G
|
|
chr1_+_103617427
Show fit
|
3.83 |
ENST00000423678.2
ENST00000414303.7
|
AMY2A
|
amylase alpha 2A
|
|
chr17_+_70075317
Show fit
|
3.75 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16
|
|
chr7_-_150323489
Show fit
|
3.75 |
ENST00000683684.1
ENST00000478393.5
|
ACTR3C
|
actin related protein 3C
|
|
chr12_-_110920568
Show fit
|
3.71 |
ENST00000548438.1
ENST00000228841.15
|
MYL2
|
myosin light chain 2
|
|
chr8_-_123737378
Show fit
|
3.60 |
ENST00000419625.6
ENST00000262219.10
|
ANXA13
|
annexin A13
|
|
chr12_-_18090141
Show fit
|
3.60 |
ENST00000536890.1
|
RERGL
|
RERG like
|
|
chr22_-_21982748
Show fit
|
3.57 |
ENST00000398793.6
ENST00000437929.5
ENST00000456075.5
ENST00000434517.1
ENST00000424393.5
ENST00000357179.10
ENST00000449704.5
ENST00000437103.1
|
TOP3B
|
DNA topoisomerase III beta
|
|
chr6_+_125749623
Show fit
|
3.50 |
ENST00000368364.4
|
HEY2
|
hes related family bHLH transcription factor with YRPW motif 2
|
|
chr19_+_41796616
Show fit
|
3.43 |
ENST00000344550.4
|
CEACAM3
|
CEA cell adhesion molecule 3
|
|
chr13_-_103066411
Show fit
|
3.43 |
ENST00000245312.5
|
SLC10A2
|
solute carrier family 10 member 2
|
|
chr22_+_30607072
Show fit
|
3.41 |
ENST00000450638.5
|
TCN2
|
transcobalamin 2
|
|
chr12_+_100473708
Show fit
|
3.41 |
ENST00000548884.5
ENST00000392986.8
|
NR1H4
|
nuclear receptor subfamily 1 group H member 4
|
|
chr19_+_41796580
Show fit
|
3.40 |
ENST00000357396.8
ENST00000630848.2
|
CEACAM3
|
CEA cell adhesion molecule 3
|
|
chr6_-_29045175
Show fit
|
3.37 |
ENST00000377175.2
|
OR2W1
|
olfactory receptor family 2 subfamily W member 1
|
|
chr10_-_70888546
Show fit
|
3.31 |
ENST00000299299.4
|
PCBD1
|
pterin-4 alpha-carbinolamine dehydratase 1
|
|
chr22_+_30607145
Show fit
|
3.24 |
ENST00000405742.7
|
TCN2
|
transcobalamin 2
|
|
chr13_+_95433593
Show fit
|
3.23 |
ENST00000376873.7
|
CLDN10
|
claudin 10
|
|
chr22_+_30607167
Show fit
|
3.21 |
ENST00000215838.8
|
TCN2
|
transcobalamin 2
|
|
chr17_+_70075215
Show fit
|
3.20 |
ENST00000283936.5
ENST00000615244.4
ENST00000392671.6
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16
|
|
chr2_-_74328166
Show fit
|
3.18 |
ENST00000358683.8
|
SLC4A5
|
solute carrier family 4 member 5
|
|
chr15_+_58138368
Show fit
|
3.16 |
ENST00000219919.9
ENST00000536493.1
|
AQP9
|
aquaporin 9
|
|
chr9_+_100099233
Show fit
|
3.08 |
ENST00000262457.7
ENST00000262456.6
ENST00000374921.3
|
INVS
|
inversin
|
|
chr7_+_130486171
Show fit
|
2.97 |
ENST00000341441.9
ENST00000416162.7
|
MEST
|
mesoderm specific transcript
|
|
chr1_-_207032749
Show fit
|
2.88 |
ENST00000359470.6
ENST00000461135.2
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr12_-_18090185
Show fit
|
2.73 |
ENST00000229002.6
ENST00000538724.6
|
RERGL
|
RERG like
|
|
chr4_-_69653223
Show fit
|
2.72 |
ENST00000286604.8
ENST00000505512.1
ENST00000514019.1
|
UGT2A1
|
UDP glucuronosyltransferase family 2 member A1 complex locus
|
|
chr7_+_130486324
Show fit
|
2.69 |
ENST00000427521.6
ENST00000378576.9
|
MEST
|
mesoderm specific transcript
|
|
chr12_+_18242955
Show fit
|
2.65 |
ENST00000676171.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma
|
|
chr6_+_167291309
Show fit
|
2.64 |
ENST00000230256.8
|
UNC93A
|
unc-93 homolog A
|
|
chr5_+_132369691
Show fit
|
2.60 |
ENST00000245407.8
|
SLC22A5
|
solute carrier family 22 member 5
|
|
chr1_+_207088825
Show fit
|
2.56 |
ENST00000367078.8
|
C4BPB
|
complement component 4 binding protein beta
|
|
chr14_+_24120956
Show fit
|
2.53 |
ENST00000558325.2
|
ENSG00000259371.2
|
novel protein
|
|
chr1_+_207089283
Show fit
|
2.50 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein beta
|
|
chr6_-_117425905
Show fit
|
2.49 |
ENST00000368507.8
|
ROS1
|
ROS proto-oncogene 1, receptor tyrosine kinase
|
|
chr6_+_106086316
Show fit
|
2.49 |
ENST00000369091.6
ENST00000369096.9
|
PRDM1
|
PR/SET domain 1
|
|
chr6_-_117425855
Show fit
|
2.38 |
ENST00000368508.7
|
ROS1
|
ROS proto-oncogene 1, receptor tyrosine kinase
|
|
chr16_-_87936529
Show fit
|
2.36 |
ENST00000649794.3
ENST00000649158.1
ENST00000648177.1
|
CA5A
|
carbonic anhydrase 5A
|
|
chr19_-_41627051
Show fit
|
2.25 |
ENST00000221954.6
ENST00000600925.1
|
CEACAM4
|
CEA cell adhesion molecule 4
|
|
chr4_+_119135825
Show fit
|
2.19 |
ENST00000307128.6
|
MYOZ2
|
myozenin 2
|
|
chr4_+_168092530
Show fit
|
2.09 |
ENST00000359299.8
|
ANXA10
|
annexin A10
|
|
chr12_+_25052732
Show fit
|
2.08 |
ENST00000547044.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2
|
|
chr6_+_167122742
Show fit
|
1.97 |
ENST00000341935.9
ENST00000349984.6
|
CCR6
|
C-C motif chemokine receptor 6
|
|
chr7_-_123199960
Show fit
|
1.89 |
ENST00000194130.7
|
SLC13A1
|
solute carrier family 13 member 1
|
|
chr1_+_207089233
Show fit
|
1.87 |
ENST00000243611.9
ENST00000367076.7
|
C4BPB
|
complement component 4 binding protein beta
|
|
chr15_-_55408018
Show fit
|
1.84 |
ENST00000569205.5
|
CCPG1
|
cell cycle progression 1
|
|
chr12_-_8540873
Show fit
|
1.77 |
ENST00000545274.5
ENST00000446457.6
ENST00000299663.8
|
CLEC4E
|
C-type lectin domain family 4 member E
|
|
chr11_+_560956
Show fit
|
1.68 |
ENST00000397582.7
ENST00000397583.8
|
RASSF7
|
Ras association domain family member 7
|
|
chr3_+_38265802
Show fit
|
1.65 |
ENST00000311856.9
|
SLC22A13
|
solute carrier family 22 member 13
|
|
chr12_+_20810698
Show fit
|
1.58 |
ENST00000540853.5
ENST00000381545.8
|
SLCO1B3
|
solute carrier organic anion transporter family member 1B3
|
|
chr10_-_37857582
Show fit
|
1.47 |
ENST00000395867.8
ENST00000611278.4
|
ZNF248
|
zinc finger protein 248
|
|
chr5_-_35195236
Show fit
|
1.35 |
ENST00000509839.5
|
PRLR
|
prolactin receptor
|
|
chr15_+_85380565
Show fit
|
1.34 |
ENST00000559362.5
ENST00000394518.7
|
AKAP13
|
A-kinase anchoring protein 13
|
|
chrX_+_1591590
Show fit
|
1.31 |
ENST00000313871.9
ENST00000381261.8
|
AKAP17A
|
A-kinase anchoring protein 17A
|
|
chr8_-_107498041
Show fit
|
1.25 |
ENST00000297450.7
|
ANGPT1
|
angiopoietin 1
|
|
chr4_-_993430
Show fit
|
1.24 |
ENST00000361661.6
ENST00000622731.4
|
SLC26A1
|
solute carrier family 26 member 1
|
|
chr22_+_32043253
Show fit
|
1.20 |
ENST00000266088.9
|
SLC5A1
|
solute carrier family 5 member 1
|
|
chr20_+_44401222
Show fit
|
1.12 |
ENST00000316099.9
|
HNF4A
|
hepatocyte nuclear factor 4 alpha
|
|
chr1_-_197067234
Show fit
|
1.09 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain
|
|
chr7_-_115968302
Show fit
|
1.05 |
ENST00000457268.5
|
TFEC
|
transcription factor EC
|
|
chr10_-_99930893
Show fit
|
1.05 |
ENST00000636706.1
|
DNMBP
|
dynamin binding protein
|
|
chr11_-_83682385
Show fit
|
1.04 |
ENST00000426717.6
|
DLG2
|
discs large MAGUK scaffold protein 2
|
|
chr8_-_107497909
Show fit
|
0.98 |
ENST00000517746.6
|
ANGPT1
|
angiopoietin 1
|
|
chr3_-_192727500
Show fit
|
0.97 |
ENST00000430714.5
ENST00000418610.1
ENST00000445105.7
ENST00000684728.1
ENST00000448795.5
ENST00000683935.1
ENST00000684282.1
ENST00000683451.1
|
FGF12
|
fibroblast growth factor 12
|
|
chr1_-_113887574
Show fit
|
0.92 |
ENST00000393316.8
|
BCL2L15
|
BCL2 like 15
|
|
chr11_-_83682414
Show fit
|
0.91 |
ENST00000404783.7
|
DLG2
|
discs large MAGUK scaffold protein 2
|