Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
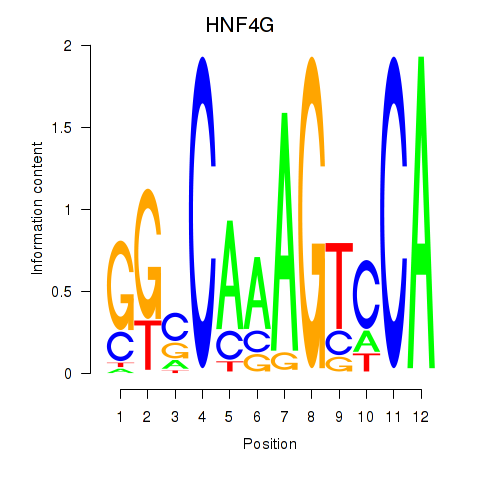
Results for HNF4G
Z-value: 3.83
Transcription factors associated with HNF4G
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF4G
|
ENSG00000164749.13 | HNF4G |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HNF4G | hg38_v1_chr8_+_75539862_75539892, hg38_v1_chr8_+_75539893_75539968 | -0.29 | 1.7e-05 | Click! |
Activity profile of HNF4G motif
Sorted Z-values of HNF4G motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HNF4G
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.1 | 40.5 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 4.6 | 13.7 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 4.5 | 13.5 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 4.0 | 36.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 3.3 | 13.3 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 3.3 | 13.3 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 3.2 | 19.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 3.0 | 9.0 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 3.0 | 8.9 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 2.9 | 11.8 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 2.6 | 34.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 2.6 | 7.7 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 2.3 | 18.4 | GO:0046061 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 2.3 | 9.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 2.3 | 38.7 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 2.3 | 6.8 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 2.1 | 6.4 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 2.0 | 5.9 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 1.9 | 13.5 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 1.9 | 19.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 1.8 | 1.8 | GO:0033076 | isoquinoline alkaloid metabolic process(GO:0033076) |
| 1.6 | 9.6 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 1.5 | 4.6 | GO:0002445 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 1.5 | 7.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 1.4 | 8.4 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 1.4 | 5.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.3 | 5.4 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 1.3 | 4.0 | GO:0005999 | xylulose biosynthetic process(GO:0005999) |
| 1.3 | 5.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 1.3 | 1.3 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 1.2 | 3.7 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 1.2 | 3.7 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 1.2 | 3.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 1.2 | 7.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 1.1 | 21.7 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 1.1 | 4.5 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 1.0 | 5.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 1.0 | 14.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.0 | 3.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 1.0 | 3.0 | GO:0042946 | glucoside transport(GO:0042946) |
| 1.0 | 20.0 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 1.0 | 3.9 | GO:0030047 | actin modification(GO:0030047) |
| 1.0 | 3.8 | GO:0002159 | desmosome assembly(GO:0002159) |
| 1.0 | 2.9 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.9 | 7.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.9 | 2.8 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) trochlear nerve development(GO:0021558) regulation of timing of neuron differentiation(GO:0060164) |
| 0.9 | 10.9 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.9 | 4.4 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.9 | 6.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.8 | 6.5 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.7 | 7.8 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.7 | 11.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.7 | 2.7 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.7 | 24.8 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.6 | 18.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.6 | 3.1 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.6 | 14.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.6 | 9.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.6 | 4.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.6 | 1.8 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.6 | 5.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.6 | 4.0 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.6 | 3.4 | GO:0060313 | negative regulation of blood vessel remodeling(GO:0060313) |
| 0.6 | 1.7 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.5 | 2.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.5 | 7.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.5 | 7.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.5 | 1.5 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.5 | 7.0 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.5 | 13.0 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.5 | 4.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.5 | 8.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.5 | 4.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.5 | 9.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 1.3 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.4 | 2.1 | GO:0098706 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.4 | 2.4 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.4 | 5.2 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.4 | 11.8 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.4 | 1.1 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.4 | 9.7 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.4 | 3.0 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.4 | 2.9 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.4 | 14.3 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.3 | 2.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.3 | 3.4 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.3 | 2.7 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 1.0 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.3 | 2.7 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 3.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 8.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.3 | 5.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 2.9 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.3 | 8.5 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.3 | 2.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) positive regulation of autophagosome maturation(GO:1901098) |
| 0.3 | 2.7 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.3 | 3.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.3 | 1.8 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) negative regulation of lung blood pressure(GO:0061767) |
| 0.3 | 1.2 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.3 | 6.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.3 | 4.5 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.3 | 0.8 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.3 | 10.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.3 | 0.8 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.3 | 0.8 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.3 | 16.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.3 | 0.8 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.3 | 9.5 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.3 | 4.6 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.3 | 6.7 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 0.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.3 | 20.0 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.3 | 3.4 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.3 | 1.8 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.3 | 1.8 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.3 | 1.5 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.2 | 6.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.2 | 1.7 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 0.5 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.2 | 1.5 | GO:0007044 | cell-substrate junction assembly(GO:0007044) |
| 0.2 | 2.9 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.2 | 7.0 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.2 | 3.3 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.2 | 0.9 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.2 | 4.4 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.2 | 0.6 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.2 | 2.5 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.2 | 1.2 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.2 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 4.7 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.2 | 2.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 0.6 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 | 1.9 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.2 | 30.7 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.2 | 1.1 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.2 | 0.4 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.2 | 1.2 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 1.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.2 | 1.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 3.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 4.2 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.2 | 1.5 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.2 | 0.7 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 11.7 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 0.6 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.2 | 4.1 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.2 | 12.2 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.2 | 2.9 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 0.7 | GO:1902998 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 1.2 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 3.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 1.6 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 3.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 5.6 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 2.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 2.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 6.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 3.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 5.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 1.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 1.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.8 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.7 | GO:0043316 | cytotoxic T cell degranulation(GO:0043316) positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 1.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.8 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 2.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 3.7 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 2.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.5 | GO:0038170 | hormone-mediated apoptotic signaling pathway(GO:0008628) somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 4.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 8.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 1.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.4 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) |
| 0.1 | 0.4 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 5.6 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 3.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 3.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 5.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.8 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.1 | 0.2 | GO:0009804 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.1 | 2.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 2.9 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.1 | 2.0 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 3.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 4.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 3.4 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.1 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.7 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 3.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 2.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 11.9 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 5.8 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 4.0 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 6.6 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.2 | GO:0035811 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) negative regulation of urine volume(GO:0035811) |
| 0.0 | 2.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 2.7 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 2.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.5 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.7 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 1.0 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 1.1 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 1.8 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.8 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.3 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 5.1 | GO:0032886 | regulation of microtubule-based process(GO:0032886) |
| 0.0 | 0.7 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.5 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 1.2 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 1.2 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 1.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 2.6 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 3.3 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.0 | 2.4 | GO:0009108 | coenzyme biosynthetic process(GO:0009108) |
| 0.0 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0098838 | methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 1.9 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.3 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 2.7 | 21.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 2.5 | 10.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 2.0 | 10.2 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 2.0 | 20.0 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 1.9 | 13.5 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.8 | 9.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.7 | 6.7 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 1.6 | 11.5 | GO:0032021 | NELF complex(GO:0032021) |
| 1.5 | 24.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 1.4 | 5.4 | GO:0071942 | XPC complex(GO:0071942) |
| 1.2 | 3.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.1 | 7.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 1.1 | 6.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.9 | 8.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.9 | 5.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.8 | 5.9 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.8 | 9.8 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.7 | 4.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.7 | 11.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.6 | 4.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.6 | 8.9 | GO:0090543 | Flemming body(GO:0090543) |
| 0.6 | 45.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.6 | 2.9 | GO:0031523 | Myb complex(GO:0031523) |
| 0.6 | 9.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.5 | 7.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.5 | 2.7 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.5 | 5.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.5 | 31.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.5 | 53.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.5 | 4.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.5 | 9.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.4 | 5.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 3.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 11.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.4 | 4.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 0.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 1.8 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 8.0 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.3 | 10.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 11.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.3 | 8.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.3 | 3.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 19.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.3 | 2.5 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 1.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.2 | 31.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 8.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 4.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 12.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 0.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 1.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 2.4 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.2 | 2.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 1.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 2.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 2.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 0.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 9.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 1.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 2.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 4.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 3.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 15.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 4.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 5.7 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 3.4 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 2.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 2.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.8 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 11.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 9.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 5.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 27.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 3.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 31.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 2.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 4.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 5.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 7.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 15.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 1.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 3.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.5 | GO:0038201 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.1 | 0.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 2.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 2.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.6 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 129.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 2.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 29.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 1.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 2.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 12.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 5.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.3 | 34.0 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 9.6 | 28.8 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 6.1 | 18.4 | GO:0044713 | GTP diphosphatase activity(GO:0036219) 2-hydroxy-adenosine triphosphate pyrophosphatase activity(GO:0044713) 2-hydroxy-(deoxy)adenosine-triphosphate pyrophosphatase activity(GO:0044714) ATP diphosphatase activity(GO:0047693) |
| 5.8 | 11.7 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 4.8 | 19.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 4.5 | 13.5 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 4.3 | 13.0 | GO:0047012 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) sterol-4-alpha-carboxylate 3-dehydrogenase (decarboxylating) activity(GO:0047012) |
| 3.1 | 9.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 2.5 | 10.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 2.4 | 7.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 2.3 | 6.8 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 2.2 | 20.0 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 2.1 | 8.5 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 2.1 | 43.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 2.0 | 7.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 1.9 | 13.3 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 1.9 | 7.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.7 | 6.9 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 1.7 | 11.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 1.7 | 6.7 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 1.7 | 6.7 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.5 | 7.7 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 1.5 | 24.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 1.4 | 7.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.1 | 5.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.1 | 5.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 1.1 | 9.6 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 1.1 | 4.2 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 1.0 | 3.0 | GO:0042947 | glucoside transmembrane transporter activity(GO:0042947) |
| 1.0 | 18.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 1.0 | 43.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 1.0 | 8.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.0 | 3.0 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 1.0 | 8.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.0 | 3.0 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 1.0 | 11.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.0 | 20.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.8 | 4.0 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.8 | 4.0 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.7 | 5.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.7 | 2.6 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.6 | 0.6 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.6 | 6.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.6 | 17.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.6 | 2.9 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.6 | 4.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.5 | 2.7 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.5 | 5.9 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.5 | 2.7 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.5 | 2.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.5 | 2.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.5 | 3.6 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.5 | 3.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.5 | 13.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.5 | 3.3 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.4 | 11.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.4 | 1.3 | GO:0052852 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.4 | 34.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.4 | 9.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 3.8 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.4 | 4.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.4 | 2.4 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.4 | 1.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.4 | 2.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.4 | 1.8 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.4 | 10.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.4 | 1.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.3 | 1.7 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.3 | 9.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.3 | 1.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 6.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.3 | 3.4 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.3 | 1.8 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 1.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 15.2 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.3 | 3.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 4.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 7.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.3 | 9.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 6.0 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.3 | 5.8 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.3 | 3.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 1.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.3 | 9.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 1.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 1.5 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 0.7 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.2 | 5.6 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 4.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 4.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 2.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 4.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.2 | 0.7 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.2 | 1.8 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 4.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.2 | 1.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 18.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 1.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.2 | 2.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 4.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 5.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 1.2 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.2 | 6.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 0.8 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.2 | 30.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 1.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 11.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 5.0 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.2 | 5.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 2.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.6 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 1.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 4.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 1.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 1.1 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.8 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 23.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 2.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 4.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 0.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 2.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 2.0 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 1.1 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.4 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 2.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 8.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 3.7 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.1 | 4.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 12.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 0.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 2.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 12.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 1.5 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 3.6 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.1 | 13.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 2.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.6 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 2.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 4.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 4.0 | GO:0016597 | amino acid binding(GO:0016597) |
| 0.0 | 1.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 6.3 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 2.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 5.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 3.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 12.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 2.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 9.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.4 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.0 | GO:0032947 | protein complex scaffold(GO:0032947) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 59.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 13.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.4 | 9.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.3 | 29.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 5.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 7.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 17.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 7.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 5.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 4.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 11.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 6.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 10.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.2 | 7.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 12.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 9.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 5.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 3.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 11.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 3.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 3.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 3.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 4.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 4.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.9 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 4.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 4.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 13.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 3.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 5.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 43.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 1.4 | 37.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.9 | 19.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.9 | 18.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.7 | 41.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.7 | 45.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.7 | 9.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.6 | 11.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.5 | 16.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 19.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.5 | 10.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.5 | 19.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.5 | 11.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 4.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.4 | 6.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.4 | 11.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.4 | 4.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 6.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.3 | 6.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 4.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 2.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.3 | 5.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.3 | 16.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.3 | 3.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 6.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.3 | 18.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.3 | 5.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.3 | 8.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 16.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.3 | 2.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 36.5 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.2 | 11.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 15.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 7.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 3.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 3.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 3.4 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.2 | 5.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 3.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 6.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.2 | 16.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 5.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 11.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 3.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 4.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 2.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 2.0 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 2.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 5.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 6.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 3.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 12.3 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 2.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 7.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 19.8 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.1 | 1.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 7.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 3.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 2.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 3.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 2.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |