Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
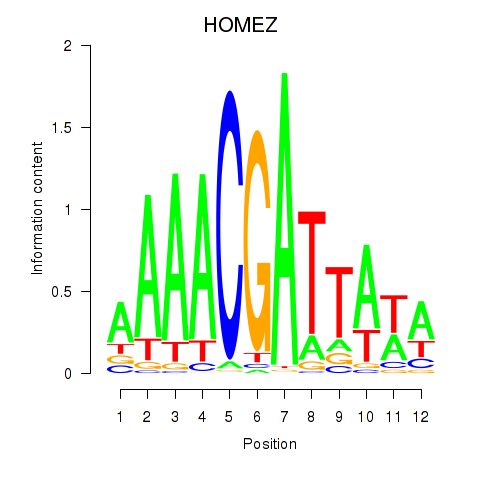
Results for HOMEZ
Z-value: 7.67
Transcription factors associated with HOMEZ
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOMEZ
|
ENSG00000215271.9 | HOMEZ |
Activity profile of HOMEZ motif
Sorted Z-values of HOMEZ motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOMEZ
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.3 | 40.0 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 7.0 | 28.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 5.6 | 16.7 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 5.0 | 20.0 | GO:2000426 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 4.8 | 14.4 | GO:1901389 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 4.2 | 33.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 4.1 | 16.2 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 3.9 | 11.6 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 3.7 | 11.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 3.7 | 7.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 3.4 | 27.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 3.4 | 13.5 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 2.9 | 17.4 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 2.8 | 8.4 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 2.8 | 8.3 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 2.8 | 11.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 2.7 | 8.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 2.7 | 10.6 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 2.5 | 27.5 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 2.4 | 7.1 | GO:0006311 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) |
| 2.3 | 9.3 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 2.3 | 34.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 2.2 | 6.5 | GO:0051793 | medium-chain fatty acid catabolic process(GO:0051793) |
| 2.1 | 19.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 2.0 | 10.0 | GO:0030242 | pexophagy(GO:0030242) |
| 2.0 | 7.9 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 1.9 | 9.7 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 1.9 | 9.6 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 1.9 | 22.8 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.9 | 20.8 | GO:0042262 | DNA protection(GO:0042262) |
| 1.8 | 7.3 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 1.8 | 5.3 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) dGDP metabolic process(GO:0046066) |
| 1.7 | 25.5 | GO:0006983 | ER overload response(GO:0006983) |
| 1.7 | 5.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 1.6 | 11.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 1.6 | 57.1 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
| 1.6 | 4.7 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 1.5 | 6.2 | GO:0050923 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 1.5 | 7.6 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 1.5 | 19.7 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 1.5 | 10.4 | GO:0051026 | chiasma assembly(GO:0051026) |
| 1.5 | 4.4 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 1.5 | 10.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 1.4 | 13.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 1.4 | 20.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 1.4 | 13.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 1.4 | 5.7 | GO:0030047 | actin modification(GO:0030047) |
| 1.4 | 4.2 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 1.4 | 5.6 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 1.4 | 6.8 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 1.3 | 61.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 1.3 | 13.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.3 | 26.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.2 | 16.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 1.2 | 7.3 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 1.2 | 3.6 | GO:2000397 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) ubiquitin-dependent endocytosis(GO:0070086) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 1.2 | 4.8 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 1.2 | 3.6 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 1.2 | 8.3 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 1.2 | 8.2 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) positive regulation of necrotic cell death(GO:0010940) |
| 1.2 | 4.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.1 | 12.5 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 1.1 | 4.5 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 1.1 | 5.4 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 1.1 | 5.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 1.0 | 10.5 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 1.0 | 4.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 1.0 | 5.9 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.9 | 7.5 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.9 | 2.7 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.9 | 5.4 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.9 | 6.9 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.8 | 4.2 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.8 | 12.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.7 | 9.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.7 | 11.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.7 | 8.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.7 | 2.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.7 | 3.5 | GO:0097021 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.7 | 5.5 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.7 | 2.0 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.7 | 6.7 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.7 | 6.7 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.7 | 2.7 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.7 | 2.6 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.6 | 3.9 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.6 | 11.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.6 | 7.2 | GO:2001138 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.6 | 4.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.6 | 5.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.6 | 1.2 | GO:1900244 | regulation of synaptic vesicle endocytosis(GO:1900242) positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.6 | 38.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.6 | 10.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.6 | 7.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.6 | 4.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.5 | 2.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.5 | 6.6 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.5 | 6.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.5 | 2.5 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.5 | 3.4 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.5 | 6.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.5 | 10.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.4 | 7.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.4 | 33.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.4 | 2.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.4 | 10.6 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.4 | 5.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.4 | 2.5 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) negative regulation of establishment of protein localization to mitochondrion(GO:1903748) regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.4 | 1.3 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.4 | 7.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 3.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.4 | 1.5 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.4 | 1.9 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.4 | 5.5 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.4 | 4.8 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.4 | 1.8 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.4 | 27.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.3 | 14.4 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.3 | 5.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.3 | 3.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.3 | 1.0 | GO:1902232 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) regulation of positive thymic T cell selection(GO:1902232) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.3 | 1.6 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.3 | 9.6 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.3 | 1.8 | GO:0060356 | leucine import(GO:0060356) |
| 0.3 | 8.3 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.3 | 1.2 | GO:0032597 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.3 | 5.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 6.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.3 | 1.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.3 | 32.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.3 | 1.4 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.3 | 6.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.3 | 1.6 | GO:0090625 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 5.4 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.3 | 22.1 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.3 | 1.8 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 7.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.3 | 1.5 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.2 | 1.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 13.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.2 | 8.2 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 1.6 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 0.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.9 | GO:0030821 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.2 | 3.0 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 10.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.2 | 6.8 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.2 | 10.6 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.2 | 8.7 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.2 | 1.3 | GO:0051958 | methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.2 | 2.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.2 | 1.6 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 | 2.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.2 | 1.0 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.2 | 1.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 9.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 1.0 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 7.4 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.2 | 3.2 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.2 | 7.6 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.2 | 9.8 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.2 | 13.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 10.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 1.9 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 1.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 13.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.2 | 0.3 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 7.4 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 4.8 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.1 | 11.0 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 0.6 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.6 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 | 0.6 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 2.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 6.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 7.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 17.1 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.1 | 0.9 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 4.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.6 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.1 | GO:0072387 | flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.1 | 1.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 3.8 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.1 | 0.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 2.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 6.5 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 1.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 2.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 1.9 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 1.7 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 2.3 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 4.1 | GO:0048208 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 9.3 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 1.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 10.1 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.1 | 4.0 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 7.3 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 2.7 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 11.3 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.1 | 7.1 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 8.4 | GO:0090002 | establishment of protein localization to plasma membrane(GO:0090002) |
| 0.0 | 1.5 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 3.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 6.4 | GO:0070585 | protein localization to mitochondrion(GO:0070585) |
| 0.0 | 5.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.9 | GO:0002003 | regulation of angiotensin levels in blood(GO:0002002) angiotensin maturation(GO:0002003) regulation of angiotensin metabolic process(GO:0060177) |
| 0.0 | 0.4 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 3.1 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 2.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 2.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.4 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.5 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 2.3 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.6 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 3.8 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 3.1 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.4 | GO:1990913 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 3.4 | 55.1 | GO:0001741 | XY body(GO:0001741) |
| 3.3 | 10.0 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 2.7 | 16.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 2.6 | 10.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 2.5 | 17.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 2.2 | 28.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 2.1 | 10.6 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 2.1 | 6.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 2.1 | 36.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 1.9 | 20.6 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 1.7 | 6.9 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 1.7 | 8.4 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 1.6 | 11.4 | GO:0031415 | NatA complex(GO:0031415) |
| 1.6 | 13.0 | GO:0000243 | commitment complex(GO:0000243) |
| 1.6 | 9.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.6 | 12.5 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 1.5 | 7.7 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 1.5 | 17.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.4 | 7.1 | GO:0032301 | MutSalpha complex(GO:0032301) MutSbeta complex(GO:0032302) |
| 1.4 | 11.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 1.3 | 6.5 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 1.3 | 20.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 1.2 | 14.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 1.2 | 4.7 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 1.1 | 13.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 1.0 | 9.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 1.0 | 11.6 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.9 | 7.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.9 | 10.6 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.9 | 24.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.8 | 7.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.8 | 3.9 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.7 | 9.6 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.7 | 3.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.7 | 17.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.7 | 3.6 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.7 | 2.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.7 | 7.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.6 | 21.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.6 | 4.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.6 | 67.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.6 | 6.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.6 | 5.9 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.6 | 16.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.6 | 4.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.6 | 4.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.5 | 10.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.5 | 10.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.5 | 8.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.5 | 5.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.5 | 3.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 7.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.4 | 1.7 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.4 | 17.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.4 | 29.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.4 | 3.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.4 | 5.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.4 | 2.5 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.3 | 25.7 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.3 | 36.3 | GO:0005901 | caveola(GO:0005901) |
| 0.3 | 5.5 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.3 | 7.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 4.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.3 | 7.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.3 | 5.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 11.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 5.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.2 | 11.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 52.5 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.2 | 4.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 4.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 25.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.2 | 11.2 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 7.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 6.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 8.6 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 29.2 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 0.5 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.2 | 3.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 7.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 15.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.2 | 6.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 4.7 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.2 | 30.5 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 47.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 2.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 6.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 20.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 7.7 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 9.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 9.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 5.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 3.3 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 6.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 9.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 5.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 4.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 3.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 20.7 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 4.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 9.1 | GO:0045121 | membrane raft(GO:0045121) |
| 0.0 | 2.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 2.7 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 1.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 5.2 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 8.4 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 4.4 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.0 | 40.0 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 4.8 | 19.3 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 4.8 | 14.4 | GO:0002135 | CTP binding(GO:0002135) |
| 4.4 | 17.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 4.3 | 13.0 | GO:0000035 | acyl binding(GO:0000035) |
| 3.7 | 11.0 | GO:0031208 | POZ domain binding(GO:0031208) |
| 3.1 | 24.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 2.9 | 20.0 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 2.4 | 7.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 2.4 | 9.6 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 2.4 | 7.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 2.4 | 18.9 | GO:0070990 | snRNP binding(GO:0070990) |
| 2.3 | 9.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 2.2 | 6.5 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 2.1 | 33.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 2.1 | 10.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 2.0 | 8.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 1.8 | 12.9 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 1.8 | 5.5 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 1.8 | 5.4 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 1.8 | 10.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.7 | 6.9 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 1.7 | 8.6 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 1.7 | 6.8 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 1.7 | 6.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 1.7 | 8.3 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 1.7 | 11.6 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 1.6 | 16.2 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.6 | 4.7 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 1.6 | 4.7 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 1.5 | 13.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 1.4 | 7.1 | GO:0032181 | double-strand/single-strand DNA junction binding(GO:0000406) single thymine insertion binding(GO:0032143) dinucleotide repeat insertion binding(GO:0032181) |
| 1.4 | 30.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 1.4 | 61.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 1.3 | 10.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.2 | 11.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 1.1 | 7.9 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 1.1 | 32.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 1.1 | 4.5 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 1.1 | 9.8 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 1.1 | 4.3 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 1.0 | 11.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.0 | 4.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.0 | 8.0 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 1.0 | 7.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.9 | 8.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.9 | 2.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.9 | 7.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.9 | 5.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.9 | 16.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.9 | 4.4 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.8 | 11.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.8 | 8.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.8 | 12.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.8 | 6.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.8 | 3.1 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.8 | 6.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.7 | 7.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.7 | 4.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.7 | 20.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.7 | 7.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.6 | 51.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.6 | 42.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.6 | 30.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.6 | 2.8 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.5 | 7.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.5 | 7.5 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.5 | 10.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.5 | 6.5 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.5 | 1.0 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.5 | 13.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.4 | 4.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.4 | 9.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.4 | 2.7 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.4 | 5.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.4 | 3.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.4 | 9.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 6.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.4 | 72.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 2.7 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.3 | 2.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.3 | 5.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 0.9 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.3 | 5.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 1.5 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.3 | 6.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 10.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 5.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 7.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 1.0 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.3 | 11.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 1.8 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 5.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 4.9 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 5.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 2.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 4.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.2 | 9.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.3 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.2 | 1.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.2 | 12.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 4.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 10.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 6.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 2.7 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 1.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.2 | 6.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 6.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 12.1 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.2 | 0.5 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.2 | 14.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 4.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 7.9 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.2 | 3.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.4 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 1.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 2.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 3.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 12.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 4.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 3.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 5.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.3 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 6.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 4.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 2.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 10.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.6 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 13.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 2.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 3.5 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 3.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.4 | GO:0022821 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 14.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 7.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 6.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.4 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 2.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.4 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 85.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.8 | 31.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.6 | 20.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.6 | 32.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.5 | 18.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.4 | 15.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 10.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 39.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.3 | 17.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 4.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 8.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 16.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 18.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 7.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 9.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.2 | 9.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 15.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 2.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 8.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 10.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 7.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 3.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 4.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 4.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 6.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 9.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 8.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 5.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 4.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 41.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 2.6 | 39.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 2.3 | 37.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 1.4 | 15.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 1.2 | 18.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 1.1 | 22.8 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.9 | 32.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.9 | 40.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.9 | 32.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.8 | 14.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.7 | 10.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.7 | 56.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.6 | 9.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.6 | 8.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.5 | 3.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.5 | 11.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.5 | 11.1 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.4 | 8.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.4 | 9.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.4 | 12.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.4 | 13.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.4 | 6.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.4 | 10.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 68.4 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.4 | 9.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.3 | 8.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 11.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.3 | 4.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 2.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.3 | 4.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.3 | 9.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 17.4 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.3 | 7.3 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.3 | 7.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 6.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 34.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 5.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 2.5 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.2 | 2.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.2 | 1.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.2 | 4.1 | REACTOME RNA POL II TRANSCRIPTION | Genes involved in RNA Polymerase II Transcription |
| 0.2 | 19.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 14.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.2 | 26.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 5.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 3.4 | REACTOME CELL CYCLE CHECKPOINTS | Genes involved in Cell Cycle Checkpoints |
| 0.2 | 2.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 4.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 21.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 4.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 11.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 4.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 5.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 3.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 8.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 12.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 2.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 3.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 2.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 3.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 3.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 2.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |