Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
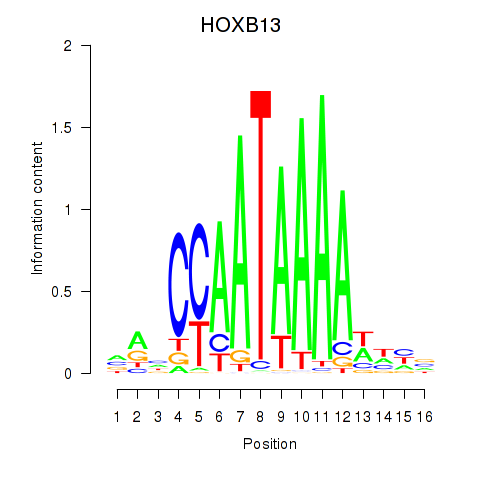
Results for HOXB13
Z-value: 0.72
Transcription factors associated with HOXB13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB13
|
ENSG00000159184.8 | HOXB13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB13 | hg38_v1_chr17_-_48728705_48728756 | -0.01 | 9.1e-01 | Click! |
Activity profile of HOXB13 motif
Sorted Z-values of HOXB13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB13
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_189179754 | 7.05 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr9_+_73151833 | 5.08 |
ENST00000456643.5
ENST00000257497.11 ENST00000415424.5 |
ANXA1
|
annexin A1 |
| chr1_+_84164962 | 4.47 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr18_-_55321986 | 4.23 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chr9_+_122370523 | 4.10 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr18_-_55322215 | 3.79 |
ENST00000457482.7
|
TCF4
|
transcription factor 4 |
| chr15_-_55249029 | 3.43 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_+_84164370 | 3.21 |
ENST00000446538.5
ENST00000610703.4 ENST00000370682.7 ENST00000394838.6 ENST00000432111.5 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr9_+_122371036 | 3.14 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr6_+_6588708 | 3.07 |
ENST00000230568.5
|
LY86
|
lymphocyte antigen 86 |
| chr1_+_65992389 | 2.32 |
ENST00000423207.6
|
PDE4B
|
phosphodiesterase 4B |
| chr18_+_3252267 | 2.32 |
ENST00000536605.1
ENST00000580887.5 |
MYL12A
|
myosin light chain 12A |
| chr2_-_24328113 | 2.15 |
ENST00000622089.4
|
ITSN2
|
intersectin 2 |
| chr11_+_118530990 | 2.05 |
ENST00000411589.6
ENST00000359862.8 ENST00000442938.6 |
TMEM25
|
transmembrane protein 25 |
| chr9_+_122371014 | 1.95 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr9_-_14180779 | 1.68 |
ENST00000380924.1
ENST00000543693.5 |
NFIB
|
nuclear factor I B |
| chr9_-_128067310 | 1.57 |
ENST00000373078.5
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr16_-_24931090 | 1.52 |
ENST00000571843.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr16_-_24930952 | 1.48 |
ENST00000571406.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr5_+_136160986 | 1.36 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr3_-_115071333 | 1.18 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_-_155910881 | 1.02 |
ENST00000609492.1
ENST00000368322.7 |
RIT1
|
Ras like without CAAX 1 |
| chr14_-_50116553 | 0.99 |
ENST00000395860.7
ENST00000395859.2 |
VCPKMT
|
valosin containing protein lysine methyltransferase |
| chr19_+_35143237 | 0.98 |
ENST00000586063.5
ENST00000270310.7 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr19_+_47994625 | 0.76 |
ENST00000339841.7
|
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr19_+_47994696 | 0.74 |
ENST00000596043.5
ENST00000597519.5 |
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr1_-_149812359 | 0.72 |
ENST00000369167.2
ENST00000545683.1 |
H2BC18
|
H2B clustered histone 18 |
| chr2_-_216013517 | 0.59 |
ENST00000263268.11
|
MREG
|
melanoregulin |
| chr8_+_96584920 | 0.59 |
ENST00000521590.5
|
SDC2
|
syndecan 2 |
| chr7_+_16661182 | 0.55 |
ENST00000446596.5
ENST00000452975.6 ENST00000438834.5 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr6_-_43053832 | 0.36 |
ENST00000265348.9
ENST00000674134.1 ENST00000674100.1 |
CUL7
|
cullin 7 |
| chr5_-_140673568 | 0.35 |
ENST00000542735.2
|
DND1
|
DND microRNA-mediated repression inhibitor 1 |
| chr4_+_67558719 | 0.32 |
ENST00000265404.7
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr4_-_163613505 | 0.32 |
ENST00000339875.9
|
MARCHF1
|
membrane associated ring-CH-type finger 1 |
| chr20_+_58907981 | 0.32 |
ENST00000656419.1
|
GNAS
|
GNAS complex locus |
| chr5_+_102808057 | 0.24 |
ENST00000684043.1
ENST00000682407.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_+_210328244 | 0.21 |
ENST00000541565.5
ENST00000413764.6 |
HHAT
|
hedgehog acyltransferase |
| chr18_+_3252208 | 0.18 |
ENST00000578562.6
|
MYL12A
|
myosin light chain 12A |
| chr1_+_78303865 | 0.16 |
ENST00000370758.5
|
PTGFR
|
prostaglandin F receptor |
| chr21_-_34526850 | 0.15 |
ENST00000481448.5
ENST00000381132.6 |
RCAN1
|
regulator of calcineurin 1 |
| chr1_+_44405164 | 0.12 |
ENST00000355387.6
|
RNF220
|
ring finger protein 220 |
| chr1_-_45491150 | 0.10 |
ENST00000372086.4
|
TESK2
|
testis associated actin remodelling kinase 2 |
| chr11_+_63838902 | 0.04 |
ENST00000377810.8
|
MARK2
|
microtubule affinity regulating kinase 2 |
| chr17_-_48728705 | 0.03 |
ENST00000290295.8
|
HOXB13
|
homeobox B13 |
| chr1_+_50105666 | 0.02 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr17_-_41382298 | 0.00 |
ENST00000394001.3
|
KRT34
|
keratin 34 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 1.4 | 7.0 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 1.3 | 7.7 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.6 | 3.4 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.4 | 9.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 3.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.2 | 1.4 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.2 | 2.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 1.7 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.3 | GO:1903973 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.1 | 0.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 1.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.2 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.1 | 2.1 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 8.0 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 2.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.4 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.2 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.6 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 1.6 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 1.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.8 | 5.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.4 | 7.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 3.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 2.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 9.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 8.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 3.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 9.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.0 | 5.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.5 | 7.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 8.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 3.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 7.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 2.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.2 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 2.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 3.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 1.0 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 7.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 5.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 8.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 5.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 3.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 8.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 7.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 9.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 2.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 4.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 3.1 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 3.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |