Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
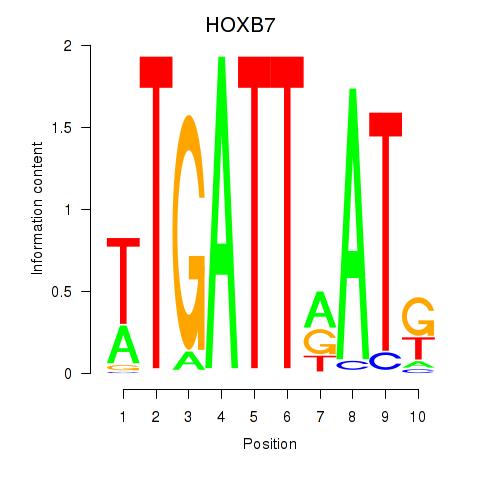
Results for HOXB7
Z-value: 5.81
Transcription factors associated with HOXB7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB7
|
ENSG00000260027.5 | HOXB7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB7 | hg38_v1_chr17_-_48610971_48611023 | -0.28 | 2.4e-05 | Click! |
Activity profile of HOXB7 motif
Sorted Z-values of HOXB7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 64.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 4.3 | 12.9 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 4.3 | 21.4 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 3.7 | 22.2 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 2.4 | 9.5 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 2.2 | 9.0 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 2.1 | 6.3 | GO:1903537 | meiotic spindle elongation(GO:0051232) meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 1.9 | 7.7 | GO:0071231 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) cellular response to folic acid(GO:0071231) |
| 1.9 | 5.6 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 1.8 | 10.9 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 1.7 | 6.9 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 1.7 | 6.9 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 1.7 | 13.4 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 1.6 | 6.5 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 1.6 | 4.8 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 1.6 | 10.9 | GO:0071279 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) cellular response to cobalt ion(GO:0071279) |
| 1.5 | 19.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.5 | 15.3 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.5 | 10.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 1.4 | 8.3 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 1.4 | 15.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.2 | 5.9 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 1.1 | 3.4 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 1.1 | 13.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 1.1 | 4.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 1.1 | 6.5 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 1.0 | 1.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.9 | 9.8 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.9 | 6.9 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.8 | 2.5 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.8 | 2.5 | GO:0039022 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.8 | 16.3 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.8 | 6.4 | GO:2000467 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.8 | 4.0 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.8 | 2.4 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.8 | 2.3 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.8 | 5.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.7 | 3.0 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.7 | 20.6 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.7 | 4.0 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.6 | 8.4 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.6 | 2.6 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.6 | 1.9 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.6 | 2.5 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.6 | 3.5 | GO:0060529 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.6 | 2.8 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.6 | 3.9 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.6 | 1.7 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.6 | 3.9 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.5 | 2.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.5 | 1.5 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.5 | 2.0 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.5 | 16.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.5 | 6.8 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.5 | 5.3 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.5 | 8.1 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.5 | 1.4 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.4 | 0.9 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.4 | 3.9 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.4 | 3.0 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.4 | 3.3 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.4 | 2.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.4 | 4.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.4 | 7.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.4 | 1.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.4 | 10.6 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.4 | 1.9 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.4 | 3.4 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.4 | 2.3 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.4 | 5.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.4 | 2.2 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.4 | 1.8 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.4 | 2.5 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.4 | 10.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.4 | 1.4 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.4 | 2.8 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.3 | 7.3 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.3 | 4.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.3 | 0.6 | GO:1902285 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) |
| 0.3 | 10.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.3 | 2.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 2.5 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.3 | 2.5 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 6.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) regulation of acrosome reaction(GO:0060046) |
| 0.3 | 2.0 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.3 | 4.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 0.8 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.3 | 0.3 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) |
| 0.3 | 3.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.3 | 2.0 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.2 | 3.7 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.2 | 3.5 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.2 | 0.7 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 1.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.2 | 2.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 12.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 20.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 4.9 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 3.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.2 | 13.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 2.5 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.2 | 13.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 1.6 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.2 | 6.0 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.2 | 2.9 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 2.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.2 | 2.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 2.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 2.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 7.2 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 1.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 3.2 | GO:0050951 | sensory perception of temperature stimulus(GO:0050951) |
| 0.1 | 0.4 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.1 | 4.2 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 22.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 6.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 7.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 2.3 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.1 | 0.8 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 2.0 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 1.8 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 2.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.2 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 1.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 2.9 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 4.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 2.4 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 1.7 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.3 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.7 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 1.7 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 2.0 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 0.4 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 1.7 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 3.2 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.1 | 0.7 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 1.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.4 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.9 | GO:0051873 | innate immune response in mucosa(GO:0002227) killing by host of symbiont cells(GO:0051873) |
| 0.1 | 2.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 1.0 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 2.0 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 1.9 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 1.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 5.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 1.8 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 1.6 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 5.7 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 6.2 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 1.5 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 2.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 2.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.3 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 1.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.8 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 4.2 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 3.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.5 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 1.2 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.0 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 8.9 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.6 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 2.1 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.5 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 4.7 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.5 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 2.4 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 2.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.0 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 | 1.7 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.0 | 0.5 | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle(GO:0007091) |
| 0.0 | 1.1 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 64.3 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.0 | 10.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.9 | 17.9 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.9 | 22.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.8 | 2.5 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.8 | 12.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.7 | 2.2 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.6 | 9.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.6 | 2.8 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.5 | 2.7 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.5 | 5.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.5 | 2.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.5 | 3.7 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.4 | 3.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.4 | 8.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 2.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 2.5 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.3 | 20.4 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 6.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 1.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.3 | 1.0 | GO:0000805 | X chromosome(GO:0000805) |
| 0.3 | 7.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 20.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.3 | 3.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 16.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 3.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 17.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 15.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 2.5 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.2 | 12.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 13.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 19.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.2 | 10.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 4.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 6.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 6.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 4.5 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 2.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 5.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 7.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 2.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 2.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 3.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 4.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.8 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 2.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 13.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 1.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 2.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 8.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 4.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 16.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 3.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.9 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.2 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 3.5 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 4.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 4.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 3.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 3.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 4.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 3.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 40.6 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 6.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 55.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 3.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.3 | GO:0035375 | zymogen binding(GO:0035375) |
| 3.2 | 9.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 3.2 | 22.2 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 2.3 | 6.9 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 2.2 | 13.2 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 1.9 | 7.7 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 1.7 | 6.8 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 1.7 | 13.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.6 | 4.8 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 1.5 | 16.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 1.3 | 5.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.2 | 7.1 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 1.1 | 9.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 1.0 | 13.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.0 | 3.0 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 1.0 | 4.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 1.0 | 3.9 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.9 | 3.7 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.9 | 11.8 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.8 | 3.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.8 | 1.6 | GO:0016160 | amylase activity(GO:0016160) |
| 0.8 | 3.9 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.8 | 2.3 | GO:0036505 | prosaposin receptor activity(GO:0036505) |
| 0.7 | 64.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.7 | 2.9 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.7 | 7.6 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.7 | 2.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.7 | 3.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.7 | 2.6 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.6 | 2.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.6 | 9.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.5 | 6.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.5 | 6.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.5 | 1.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.5 | 6.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.5 | 7.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.4 | 2.2 | GO:0047086 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.4 | 3.0 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.4 | 3.4 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.4 | 12.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 4.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.4 | 9.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.4 | 5.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.4 | 11.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 2.5 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.4 | 16.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.3 | 6.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.3 | 1.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 20.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 1.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 5.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.3 | 10.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 3.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 4.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 1.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 1.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 8.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.3 | 1.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 27.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 2.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 5.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 2.9 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.2 | 12.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 8.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 1.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.2 | 2.5 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.2 | 0.6 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.2 | 20.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 6.4 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.2 | 2.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.2 | 0.7 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.2 | 31.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.2 | 2.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 1.8 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 1.6 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.2 | 12.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.4 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 1.2 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 1.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 3.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 1.7 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 3.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 6.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 2.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 2.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.9 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 6.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.4 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 1.0 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 1.7 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 14.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 2.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 2.9 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.1 | 3.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.8 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 2.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 6.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 3.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 3.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 2.2 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 2.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 1.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 2.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.7 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 3.0 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 4.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 2.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 19.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 4.5 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.7 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 66.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 5.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 7.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 12.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 6.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 22.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 8.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.2 | 6.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 13.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 2.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 12.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 7.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 7.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 31.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 3.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 7.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 5.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 6.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 6.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 24.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 4.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 4.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 30.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 2.4 | 64.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.8 | 9.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.7 | 22.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.6 | 25.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.6 | 12.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.5 | 12.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.5 | 20.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.4 | 6.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.4 | 4.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 8.3 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.4 | 6.9 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.4 | 11.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.3 | 6.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 3.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 2.9 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.2 | 21.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 4.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 12.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 7.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 8.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 1.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 4.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 3.5 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 12.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 6.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 10.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 2.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 3.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 8.7 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.1 | 4.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 9.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 20.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 6.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 2.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 4.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 6.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 4.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 4.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.6 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 2.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 16.3 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 2.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |