Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
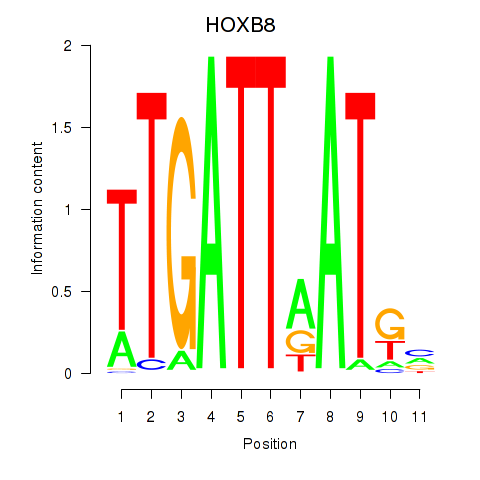
Results for HOXB8
Z-value: 2.18
Transcription factors associated with HOXB8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB8
|
ENSG00000120068.7 | HOXB8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB8 | hg38_v1_chr17_-_48613468_48613522 | 0.19 | 4.6e-03 | Click! |
Activity profile of HOXB8 motif
Sorted Z-values of HOXB8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_107824294 | 15.41 |
ENST00000369020.8
ENST00000369022.6 |
SCML4
|
Scm polycomb group protein like 4 |
| chr8_+_81280527 | 6.79 |
ENST00000297258.11
|
FABP5
|
fatty acid binding protein 5 |
| chr19_-_10587219 | 5.82 |
ENST00000591240.5
ENST00000589684.5 ENST00000591676.1 ENST00000250244.11 ENST00000590923.5 |
AP1M2
|
adaptor related protein complex 1 subunit mu 2 |
| chr8_-_80171106 | 5.78 |
ENST00000519303.6
|
TPD52
|
tumor protein D52 |
| chr14_-_106658251 | 5.36 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr3_-_111595339 | 5.36 |
ENST00000317012.5
|
ZBED2
|
zinc finger BED-type containing 2 |
| chr19_-_20565746 | 4.82 |
ENST00000594419.1
|
ZNF737
|
zinc finger protein 737 |
| chr8_+_133113483 | 4.48 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr13_+_73054969 | 4.41 |
ENST00000539231.5
|
KLF5
|
Kruppel like factor 5 |
| chr5_-_126595237 | 4.33 |
ENST00000637206.1
ENST00000553117.5 |
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1 |
| chr19_-_20565769 | 4.29 |
ENST00000427401.9
|
ZNF737
|
zinc finger protein 737 |
| chr10_+_5446601 | 4.18 |
ENST00000449083.5
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr5_+_126423363 | 3.98 |
ENST00000285689.8
|
GRAMD2B
|
GRAM domain containing 2B |
| chr8_+_97775775 | 3.88 |
ENST00000521545.7
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr3_+_148827800 | 3.86 |
ENST00000282957.9
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 |
| chr8_+_97887903 | 3.83 |
ENST00000520016.5
|
MATN2
|
matrilin 2 |
| chr2_-_55917699 | 3.74 |
ENST00000634374.1
|
EFEMP1
|
EGF containing fibulin extracellular matrix protein 1 |
| chr4_+_25160631 | 3.71 |
ENST00000510415.1
ENST00000507794.2 ENST00000512921.4 |
SEPSECS-AS1
PI4K2B
|
SEPSECS antisense RNA 1 (head to head) phosphatidylinositol 4-kinase type 2 beta |
| chr11_+_2384603 | 3.67 |
ENST00000527343.5
ENST00000464784.6 |
CD81
|
CD81 molecule |
| chr4_-_71784046 | 3.66 |
ENST00000513476.5
ENST00000273951.13 |
GC
|
GC vitamin D binding protein |
| chr4_-_70666492 | 3.52 |
ENST00000254801.9
ENST00000391614.7 |
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr11_-_102530738 | 3.44 |
ENST00000260227.5
|
MMP7
|
matrix metallopeptidase 7 |
| chr4_+_68815991 | 3.39 |
ENST00000265403.12
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase family 2 member B10 |
| chr1_+_236394268 | 3.33 |
ENST00000334232.9
|
EDARADD
|
EDAR associated death domain |
| chr8_+_127735597 | 3.29 |
ENST00000651626.1
|
MYC
|
MYC proto-oncogene, bHLH transcription factor |
| chr9_+_128149447 | 3.23 |
ENST00000277480.7
ENST00000372998.1 |
LCN2
|
lipocalin 2 |
| chr1_+_103617427 | 3.22 |
ENST00000423678.2
ENST00000414303.7 |
AMY2A
|
amylase alpha 2A |
| chr22_+_22822658 | 3.17 |
ENST00000620395.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8 |
| chr12_+_20815672 | 3.16 |
ENST00000261196.6
ENST00000381541.7 ENST00000540229.1 |
SLCO1B3
SLCO1B3-SLCO1B7
|
solute carrier organic anion transporter family member 1B3 SLCO1B3-SLCO1B7 readthrough |
| chr5_-_42825884 | 3.08 |
ENST00000506577.5
|
SELENOP
|
selenoprotein P |
| chr10_+_46375645 | 3.08 |
ENST00000622769.4
|
ANXA8L1
|
annexin A8 like 1 |
| chr16_-_20327426 | 3.05 |
ENST00000575582.5
ENST00000341642.9 ENST00000381362.8 ENST00000572347.5 ENST00000572478.5 ENST00000302555.10 |
GP2
|
glycoprotein 2 |
| chr13_-_48037934 | 3.03 |
ENST00000646804.1
ENST00000643246.1 |
SUCLA2
|
succinate-CoA ligase ADP-forming subunit beta |
| chr11_-_13495984 | 3.03 |
ENST00000282091.6
|
PTH
|
parathyroid hormone |
| chr12_+_25052634 | 3.03 |
ENST00000548766.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr10_+_46375619 | 3.02 |
ENST00000584982.7
ENST00000613703.4 |
ANXA8L1
|
annexin A8 like 1 |
| chr11_-_13496018 | 2.93 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr14_-_54441325 | 2.92 |
ENST00000556113.1
ENST00000553660.5 ENST00000216416.9 ENST00000395573.8 ENST00000557690.5 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr5_+_126423176 | 2.90 |
ENST00000542322.5
ENST00000544396.5 |
GRAMD2B
|
GRAM domain containing 2B |
| chr5_+_160421847 | 2.86 |
ENST00000352433.10
ENST00000517480.1 ENST00000520452.5 ENST00000393964.1 |
PTTG1
|
PTTG1 regulator of sister chromatid separation, securin |
| chr10_+_46375721 | 2.85 |
ENST00000616785.1
ENST00000611655.1 ENST00000619162.5 |
ENSG00000285402.1
ANXA8L1
|
novel transcript annexin A8 like 1 |
| chr3_-_15440560 | 2.85 |
ENST00000595627.5
ENST00000597949.1 ENST00000494875.3 ENST00000595975.1 ENST00000598878.1 |
EAF1-AS1
METTL6
|
EAF1 antisense RNA 1 methyltransferase like 6 |
| chr4_+_69096494 | 2.84 |
ENST00000508661.5
ENST00000622664.1 |
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7 |
| chr11_+_72192126 | 2.84 |
ENST00000393676.5
|
FOLR1
|
folate receptor alpha |
| chr4_+_69096467 | 2.82 |
ENST00000305231.12
|
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7 |
| chr6_-_99568713 | 2.78 |
ENST00000520371.5
|
CCNC
|
cyclin C |
| chr5_-_179617581 | 2.78 |
ENST00000523449.5
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr5_+_126423122 | 2.75 |
ENST00000515200.5
|
GRAMD2B
|
GRAM domain containing 2B |
| chr2_+_186590022 | 2.75 |
ENST00000261023.8
ENST00000374907.7 |
ITGAV
|
integrin subunit alpha V |
| chr4_-_69961007 | 2.65 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chr5_-_123036664 | 2.62 |
ENST00000306442.5
|
PPIC
|
peptidylprolyl isomerase C |
| chr12_+_25052732 | 2.61 |
ENST00000547044.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr4_+_69280472 | 2.59 |
ENST00000335568.10
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase family 2 member B28 |
| chr4_+_73409340 | 2.59 |
ENST00000511370.1
|
ALB
|
albumin |
| chr11_+_134331874 | 2.59 |
ENST00000339772.9
ENST00000535456.7 |
GLB1L2
|
galactosidase beta 1 like 2 |
| chr3_+_148791058 | 2.58 |
ENST00000491148.5
|
CPB1
|
carboxypeptidase B1 |
| chr22_-_30246739 | 2.56 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr21_-_42315336 | 2.56 |
ENST00000398431.2
ENST00000518498.3 |
TFF3
|
trefoil factor 3 |
| chr14_-_67412112 | 2.55 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr17_-_66229380 | 2.53 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr2_+_1414382 | 2.52 |
ENST00000423320.5
ENST00000346956.7 ENST00000382198.5 |
TPO
|
thyroid peroxidase |
| chr1_+_19596960 | 2.50 |
ENST00000617872.4
ENST00000322753.7 ENST00000602662.1 |
MICOS10
NBL1
|
mitochondrial contact site and cristae organizing system subunit 10 NBL1, DAN family BMP antagonist |
| chr9_-_21995301 | 2.50 |
ENST00000498628.6
|
CDKN2A
|
cyclin dependent kinase inhibitor 2A |
| chr21_-_10649835 | 2.40 |
ENST00000622028.1
|
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr2_+_191276885 | 2.39 |
ENST00000392316.5
|
MYO1B
|
myosin IB |
| chr2_+_233671879 | 2.39 |
ENST00000354728.5
|
UGT1A9
|
UDP glucuronosyltransferase family 1 member A9 |
| chr11_+_102112445 | 2.39 |
ENST00000524575.5
|
YAP1
|
Yes1 associated transcriptional regulator |
| chr15_-_21718245 | 2.38 |
ENST00000630556.1
|
ENSG00000281179.1
|
novel gene identicle to IGHV1OR15-1 |
| chr1_-_200620729 | 2.35 |
ENST00000367350.5
|
KIF14
|
kinesin family member 14 |
| chr16_-_20327801 | 2.34 |
ENST00000381360.9
|
GP2
|
glycoprotein 2 |
| chr18_+_657637 | 2.33 |
ENST00000323274.15
|
TYMS
|
thymidylate synthetase |
| chr9_+_33795551 | 2.32 |
ENST00000379405.4
|
PRSS3
|
serine protease 3 |
| chr12_-_91111460 | 2.32 |
ENST00000266718.5
|
LUM
|
lumican |
| chr19_-_17377334 | 2.29 |
ENST00000252590.9
ENST00000599426.1 |
PLVAP
|
plasmalemma vesicle associated protein |
| chr2_+_89884740 | 2.29 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr1_-_94925759 | 2.28 |
ENST00000415017.1
ENST00000545882.5 |
CNN3
|
calponin 3 |
| chr4_-_69214743 | 2.28 |
ENST00000446444.2
|
UGT2B11
|
UDP glucuronosyltransferase family 2 member B11 |
| chr14_-_106593319 | 2.26 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr4_+_168497066 | 2.23 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr5_+_178859924 | 2.21 |
ENST00000322434.8
|
ZNF354B
|
zinc finger protein 354B |
| chr12_-_54259531 | 2.18 |
ENST00000550411.5
ENST00000439541.6 |
CBX5
|
chromobox 5 |
| chr8_+_30387064 | 2.16 |
ENST00000523115.5
ENST00000519647.5 |
RBPMS
|
RNA binding protein, mRNA processing factor |
| chr10_-_46046264 | 2.15 |
ENST00000581478.5
ENST00000582163.3 |
MSMB
|
microseminoprotein beta |
| chr2_+_1484663 | 2.14 |
ENST00000446278.5
ENST00000469607.3 |
TPO
|
thyroid peroxidase |
| chr4_+_168497044 | 2.12 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein |
| chrX_+_120604199 | 2.12 |
ENST00000371315.3
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr8_-_47960126 | 2.11 |
ENST00000314191.7
ENST00000338368.7 |
PRKDC
|
protein kinase, DNA-activated, catalytic subunit |
| chr13_-_106568107 | 2.11 |
ENST00000400198.8
|
ARGLU1
|
arginine and glutamate rich 1 |
| chr2_+_176107272 | 2.11 |
ENST00000249504.7
|
HOXD11
|
homeobox D11 |
| chr17_+_47651061 | 2.09 |
ENST00000540627.5
|
KPNB1
|
karyopherin subunit beta 1 |
| chr15_+_41286011 | 2.09 |
ENST00000661438.1
|
ENSG00000285920.2
|
novel protein |
| chr1_+_103750406 | 2.09 |
ENST00000370079.3
|
AMY1C
|
amylase alpha 1C |
| chr7_+_16661182 | 2.07 |
ENST00000446596.5
ENST00000452975.6 ENST00000438834.5 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr5_+_126360113 | 2.06 |
ENST00000513040.5
|
GRAMD2B
|
GRAM domain containing 2B |
| chr10_+_60778331 | 2.05 |
ENST00000519078.6
ENST00000316629.8 ENST00000395284.8 |
CDK1
|
cyclin dependent kinase 1 |
| chr20_+_36214373 | 2.05 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr11_-_33717409 | 2.02 |
ENST00000651485.1
|
CD59
|
CD59 molecule (CD59 blood group) |
| chr18_+_3252208 | 2.01 |
ENST00000578562.6
|
MYL12A
|
myosin light chain 12A |
| chr18_+_7754959 | 1.99 |
ENST00000400053.8
|
PTPRM
|
protein tyrosine phosphatase receptor type M |
| chr1_-_103696209 | 1.99 |
ENST00000330330.10
|
AMY1B
|
amylase alpha 1B |
| chr2_+_90038848 | 1.97 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr13_+_29428709 | 1.97 |
ENST00000542829.1
|
MTUS2
|
microtubule associated scaffold protein 2 |
| chr10_+_60778490 | 1.95 |
ENST00000448257.6
ENST00000614696.4 |
CDK1
|
cyclin dependent kinase 1 |
| chr11_-_14499803 | 1.95 |
ENST00000439561.7
ENST00000534234.5 |
COPB1
|
COPI coat complex subunit beta 1 |
| chr6_-_131000722 | 1.94 |
ENST00000528282.5
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr1_+_53894181 | 1.92 |
ENST00000361921.8
ENST00000322679.10 ENST00000613679.4 ENST00000617230.2 ENST00000610607.4 ENST00000532493.5 ENST00000525202.5 ENST00000524406.5 ENST00000388876.3 |
DIO1
|
iodothyronine deiodinase 1 |
| chr2_+_233729042 | 1.92 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3 |
| chr11_-_14499833 | 1.91 |
ENST00000249923.7
ENST00000529866.5 |
COPB1
|
COPI coat complex subunit beta 1 |
| chr19_+_52429181 | 1.91 |
ENST00000301085.8
|
ZNF534
|
zinc finger protein 534 |
| chr8_-_90082871 | 1.89 |
ENST00000265431.7
|
CALB1
|
calbindin 1 |
| chr6_+_36243203 | 1.87 |
ENST00000312917.9
ENST00000388715.7 |
PNPLA1
|
patatin like phospholipase domain containing 1 |
| chr3_-_38794042 | 1.86 |
ENST00000643924.1
|
SCN10A
|
sodium voltage-gated channel alpha subunit 10 |
| chr16_-_70289480 | 1.85 |
ENST00000261772.13
ENST00000675953.1 ENST00000675691.1 ENST00000675133.1 ENST00000674512.1 ENST00000675045.1 ENST00000675853.1 ENST00000565361.3 ENST00000674963.1 ENST00000674691.1 |
AARS1
|
alanyl-tRNA synthetase 1 |
| chr8_+_103880412 | 1.85 |
ENST00000436393.6
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr8_+_22565655 | 1.82 |
ENST00000523965.5
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr6_+_29461440 | 1.82 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor family 2 subfamily H member 1 |
| chr14_-_106470788 | 1.81 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr4_+_74365136 | 1.80 |
ENST00000244869.3
|
EREG
|
epiregulin |
| chr15_+_84235773 | 1.80 |
ENST00000510439.7
ENST00000422563.6 |
GOLGA6L4
|
golgin A6 family like 4 |
| chr9_-_114074969 | 1.79 |
ENST00000466610.6
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr20_-_56392131 | 1.79 |
ENST00000422322.5
ENST00000371356.6 ENST00000451915.1 ENST00000347343.6 ENST00000395911.5 ENST00000395915.8 ENST00000395907.5 ENST00000441357.5 ENST00000456249.5 ENST00000420474.5 ENST00000395914.5 ENST00000312783.10 ENST00000395913.7 |
AURKA
|
aurora kinase A |
| chr7_-_32490361 | 1.78 |
ENST00000410044.5
ENST00000450169.7 ENST00000409987.5 ENST00000409782.5 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr18_+_31591869 | 1.78 |
ENST00000237014.8
|
TTR
|
transthyretin |
| chr11_-_63608542 | 1.78 |
ENST00000540943.1
|
PLAAT3
|
phospholipase A and acyltransferase 3 |
| chr2_-_89010515 | 1.77 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr18_-_14132423 | 1.76 |
ENST00000589498.5
ENST00000590202.3 |
ZNF519
|
zinc finger protein 519 |
| chr12_+_64404338 | 1.76 |
ENST00000332707.10
|
XPOT
|
exportin for tRNA |
| chr2_-_264024 | 1.74 |
ENST00000403712.6
ENST00000356150.10 ENST00000626873.2 ENST00000405430.5 |
SH3YL1
|
SH3 and SYLF domain containing 1 |
| chr10_+_116545907 | 1.73 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr14_+_61762405 | 1.73 |
ENST00000216294.5
|
SNAPC1
|
small nuclear RNA activating complex polypeptide 1 |
| chr3_+_105366877 | 1.73 |
ENST00000306107.9
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr12_+_25052512 | 1.73 |
ENST00000557489.5
ENST00000354454.7 ENST00000536173.5 |
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr14_+_19743571 | 1.73 |
ENST00000642117.2
|
OR4Q3
|
olfactory receptor family 4 subfamily Q member 3 |
| chr12_-_27971970 | 1.72 |
ENST00000395872.5
ENST00000201015.8 |
PTHLH
|
parathyroid hormone like hormone |
| chr15_-_22160868 | 1.72 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr6_-_138218491 | 1.71 |
ENST00000527246.3
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr4_+_25655822 | 1.70 |
ENST00000504570.5
ENST00000382051.8 |
SLC34A2
|
solute carrier family 34 member 2 |
| chr4_+_108004482 | 1.66 |
ENST00000626637.2
|
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr19_-_14835252 | 1.65 |
ENST00000641666.1
ENST00000642030.1 ENST00000642000.1 |
OR7C1
|
olfactory receptor family 7 subfamily C member 1 |
| chr1_+_13416351 | 1.65 |
ENST00000602960.1
|
PRAMEF20
|
PRAME family member 20 |
| chr2_+_233691607 | 1.65 |
ENST00000373424.5
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6 |
| chr1_-_149812359 | 1.65 |
ENST00000369167.2
ENST00000545683.1 |
H2BC18
|
H2B clustered histone 18 |
| chr1_+_101237009 | 1.64 |
ENST00000305352.7
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr19_-_49451793 | 1.64 |
ENST00000262265.10
|
PIH1D1
|
PIH1 domain containing 1 |
| chr5_-_179618032 | 1.64 |
ENST00000519033.5
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr3_+_149474688 | 1.64 |
ENST00000305354.5
ENST00000465758.1 |
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr12_-_11269805 | 1.63 |
ENST00000538488.2
|
PRB3
|
proline rich protein BstNI subfamily 3 |
| chr6_+_121435595 | 1.62 |
ENST00000649003.1
ENST00000282561.4 |
GJA1
|
gap junction protein alpha 1 |
| chr2_+_90082635 | 1.62 |
ENST00000483379.1
|
IGKV1D-17
|
immunoglobulin kappa variable 1D-17 |
| chr3_+_41200104 | 1.62 |
ENST00000643297.1
ENST00000450969.6 |
CTNNB1
|
catenin beta 1 |
| chr3_+_105367212 | 1.61 |
ENST00000472644.6
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr2_+_206159884 | 1.61 |
ENST00000392222.7
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr2_+_33436304 | 1.60 |
ENST00000402538.7
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr8_+_103298433 | 1.60 |
ENST00000522566.5
|
FZD6
|
frizzled class receptor 6 |
| chr12_+_21372899 | 1.59 |
ENST00000240652.8
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr11_+_57597563 | 1.59 |
ENST00000619430.2
ENST00000457869.1 ENST00000340687.10 ENST00000278407.9 ENST00000378323.8 ENST00000378324.6 ENST00000403558.1 |
SERPING1
|
serpin family G member 1 |
| chr2_+_206159580 | 1.58 |
ENST00000236957.9
ENST00000392221.5 ENST00000445505.5 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr1_+_220094086 | 1.58 |
ENST00000366922.3
|
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr19_+_32581410 | 1.58 |
ENST00000586035.1
|
PDCD5
|
programmed cell death 5 |
| chr20_+_58389197 | 1.58 |
ENST00000475243.6
ENST00000395802.7 |
VAPB
|
VAMP associated protein B and C |
| chr10_+_122560751 | 1.58 |
ENST00000338354.10
ENST00000664692.1 ENST00000653442.1 ENST00000664974.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr5_-_126595185 | 1.57 |
ENST00000510111.6
ENST00000635851.1 ENST00000637964.1 ENST00000413020.6 ENST00000637782.1 ENST00000409134.8 ENST00000637272.1 ENST00000636879.1 ENST00000636743.1 ENST00000636886.1 ENST00000509270.2 |
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1 |
| chr14_-_106211453 | 1.56 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr2_+_233636445 | 1.52 |
ENST00000344644.9
|
UGT1A10
|
UDP glucuronosyltransferase family 1 member A10 |
| chr6_+_57172290 | 1.52 |
ENST00000370693.5
|
BAG2
|
BAG cochaperone 2 |
| chr1_-_53945584 | 1.50 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr5_-_134367144 | 1.50 |
ENST00000265334.9
|
CDKL3
|
cyclin dependent kinase like 3 |
| chr22_-_28800042 | 1.49 |
ENST00000405219.7
|
XBP1
|
X-box binding protein 1 |
| chr20_-_50958520 | 1.49 |
ENST00000371588.10
ENST00000371584.9 ENST00000413082.1 ENST00000371582.8 |
DPM1
|
dolichyl-phosphate mannosyltransferase subunit 1, catalytic |
| chr17_-_8383164 | 1.49 |
ENST00000584164.6
ENST00000582556.5 ENST00000648839.1 ENST00000578812.5 ENST00000583011.6 |
RPL26
|
ribosomal protein L26 |
| chr1_-_53945567 | 1.48 |
ENST00000371378.6
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr11_+_118077067 | 1.47 |
ENST00000522307.5
ENST00000523251.5 ENST00000437212.8 ENST00000522824.5 ENST00000522151.5 |
TMPRSS4
|
transmembrane serine protease 4 |
| chr15_-_82349437 | 1.47 |
ENST00000621197.4
ENST00000610657.2 ENST00000619556.4 |
GOLGA6L10
|
golgin A6 family like 10 |
| chr2_-_89117844 | 1.47 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr6_-_150025520 | 1.46 |
ENST00000367341.6
ENST00000286380.2 |
RAET1L
|
retinoic acid early transcript 1L |
| chr10_+_92593112 | 1.46 |
ENST00000260731.5
|
KIF11
|
kinesin family member 11 |
| chr14_-_105987068 | 1.44 |
ENST00000390594.3
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr11_+_118077009 | 1.44 |
ENST00000616579.4
ENST00000534111.5 |
TMPRSS4
|
transmembrane serine protease 4 |
| chr10_+_122560679 | 1.44 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr19_+_20776292 | 1.44 |
ENST00000360204.5
ENST00000344519.10 ENST00000594534.5 |
ZNF66
|
zinc finger protein 66 |
| chr12_+_26195313 | 1.43 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr11_+_844406 | 1.42 |
ENST00000397404.5
|
TSPAN4
|
tetraspanin 4 |
| chrX_+_120604084 | 1.41 |
ENST00000371317.10
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr16_-_10559135 | 1.41 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr14_-_74493275 | 1.41 |
ENST00000541064.5
|
NPC2
|
NPC intracellular cholesterol transporter 2 |
| chr14_-_106005574 | 1.40 |
ENST00000390595.3
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr1_+_103749898 | 1.39 |
ENST00000622339.5
|
AMY1C
|
amylase alpha 1C |
| chr15_-_52295792 | 1.39 |
ENST00000261839.12
|
MYO5C
|
myosin VC |
| chr12_-_27972725 | 1.38 |
ENST00000545234.6
|
PTHLH
|
parathyroid hormone like hormone |
| chr14_-_73950393 | 1.38 |
ENST00000651776.1
|
FAM161B
|
FAM161 centrosomal protein B |
| chr21_-_29061351 | 1.37 |
ENST00000432178.5
|
CCT8
|
chaperonin containing TCP1 subunit 8 |
| chr2_+_43774033 | 1.37 |
ENST00000260605.12
ENST00000406852.7 ENST00000398823.6 ENST00000605786.5 |
DYNC2LI1
|
dynein cytoplasmic 2 light intermediate chain 1 |
| chr4_+_40197023 | 1.37 |
ENST00000381799.10
|
RHOH
|
ras homolog family member H |
| chr22_+_22747383 | 1.36 |
ENST00000390311.3
|
IGLV3-16
|
immunoglobulin lambda variable 3-16 |
| chr2_-_150487658 | 1.35 |
ENST00000375734.6
ENST00000263895.9 ENST00000454202.5 |
RND3
|
Rho family GTPase 3 |
| chr1_+_165895583 | 1.35 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr22_+_44752552 | 1.35 |
ENST00000389774.6
ENST00000356099.11 ENST00000396119.6 ENST00000336963.8 ENST00000412433.5 |
ARHGAP8
|
Rho GTPase activating protein 8 |
| chr14_-_106025628 | 1.35 |
ENST00000631943.1
|
IGHV7-4-1
|
immunoglobulin heavy variable 7-4-1 |
| chr4_-_154612635 | 1.33 |
ENST00000407946.5
ENST00000405164.5 ENST00000336098.8 ENST00000393846.6 ENST00000404648.7 ENST00000443553.5 |
FGG
|
fibrinogen gamma chain |
| chr14_-_49620594 | 1.32 |
ENST00000298289.7
|
RPL36AL
|
ribosomal protein L36a like |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 1.5 | 6.0 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 1.4 | 4.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 1.1 | 4.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 1.0 | 3.0 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.9 | 2.8 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.9 | 2.8 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.9 | 2.8 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.9 | 2.6 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.9 | 2.6 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.8 | 10.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.8 | 3.3 | GO:0072185 | metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.7 | 3.7 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.7 | 6.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.6 | 3.2 | GO:0015688 | response to mycotoxin(GO:0010046) iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.6 | 2.5 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.6 | 3.7 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.6 | 2.5 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.6 | 1.8 | GO:1900195 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 0.6 | 2.3 | GO:0019860 | uracil metabolic process(GO:0019860) |
| 0.6 | 4.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.6 | 4.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.5 | 1.6 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.5 | 1.6 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.5 | 1.6 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.5 | 2.6 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.5 | 4.0 | GO:0090166 | regulation of Schwann cell differentiation(GO:0014038) Golgi disassembly(GO:0090166) |
| 0.5 | 3.2 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.5 | 1.4 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.4 | 1.8 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.4 | 3.8 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.4 | 2.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.4 | 1.7 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.4 | 1.6 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.4 | 1.6 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.4 | 2.4 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.4 | 3.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.4 | 4.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.4 | 2.5 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.4 | 2.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.3 | 3.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.3 | 3.5 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.3 | 4.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.3 | 1.7 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.3 | 7.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.3 | 1.3 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.3 | 1.0 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.3 | 1.0 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.3 | 1.6 | GO:1904501 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.3 | 1.0 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.3 | 1.0 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.3 | 1.5 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.3 | 1.2 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.3 | 2.0 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.3 | 2.8 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.3 | 2.8 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.3 | 0.8 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.3 | 1.6 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.3 | 1.8 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 0.7 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.2 | 0.5 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.2 | 1.0 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 1.7 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 0.7 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.2 | 1.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 0.7 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.2 | 1.9 | GO:0072221 | metanephric part of ureteric bud development(GO:0035502) distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.2 | 4.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 0.7 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.2 | 2.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.2 | 0.6 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.2 | 1.9 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 1.5 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 1.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 1.8 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 0.6 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.2 | 28.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 0.6 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.2 | 0.6 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 0.7 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.2 | 0.7 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 2.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.2 | 2.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.2 | 0.7 | GO:0072683 | T cell extravasation(GO:0072683) |
| 0.2 | 2.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 0.5 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.2 | 1.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 1.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 5.3 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.2 | 3.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 1.8 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.2 | 1.6 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 0.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 7.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 2.8 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 2.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 0.8 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 0.2 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.2 | 1.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.2 | 3.6 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 1.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.1 | 0.4 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 1.3 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.1 | 0.9 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.1 | 2.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.3 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.1 | 1.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 1.4 | GO:0003093 | regulation of glomerular filtration(GO:0003093) caveola assembly(GO:0070836) |
| 0.1 | 1.2 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.1 | 2.0 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.9 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.9 | GO:0046985 | negative regulation of megakaryocyte differentiation(GO:0045653) positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.4 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.1 | 3.4 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.1 | 0.5 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.4 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 1.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 5.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 4.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.6 | GO:0034372 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 0.2 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.1 | 1.3 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.5 | GO:0001079 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) ammonia assimilation cycle(GO:0019676) regulation of urea metabolic process(GO:0034255) bile acid signaling pathway(GO:0038183) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 | 0.9 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.1 | 1.2 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.1 | 2.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.6 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 1.9 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.4 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.6 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.7 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 2.8 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.4 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.1 | 0.8 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 2.0 | GO:0086027 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.1 | 1.4 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 0.9 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.8 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.9 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.5 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.3 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.1 | 0.7 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 1.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.3 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.2 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 1.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 0.3 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.9 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 1.8 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.1 | 0.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.2 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.1 | 1.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 1.2 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 3.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.8 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.8 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.5 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.1 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.1 | 0.3 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.7 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.1 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.5 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.2 | GO:0046081 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.1 | 0.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 3.0 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.1 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 0.9 | GO:0007512 | adult heart development(GO:0007512) |
| 0.1 | 6.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.3 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 3.8 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.1 | 0.6 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 1.3 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.1 | 0.4 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 1.3 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.1 | 0.8 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.3 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 | 0.6 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.8 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 1.7 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 3.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.3 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 3.8 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.6 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.1 | 0.9 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 0.2 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.1 | 1.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.2 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.1 | 0.2 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 | 0.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 2.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.0 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.3 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.9 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.7 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 1.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 6.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 1.8 | GO:0051031 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.1 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 1.0 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 2.6 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 | 0.2 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 1.3 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.8 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 4.4 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 1.3 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 1.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.4 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 1.2 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.5 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 1.0 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 3.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 3.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.6 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.7 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 3.8 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.6 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 2.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.7 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 1.6 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.9 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.7 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.4 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 0.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.3 | GO:0014904 | myotube cell development(GO:0014904) |
| 0.0 | 0.0 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.0 | 0.3 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.3 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.3 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.2 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.0 | 0.5 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.4 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 1.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.4 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.6 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 | 1.4 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 0.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 0.8 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.6 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.0 | GO:0015819 | lysine transport(GO:0015819) lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine transport(GO:1902022) L-lysine import into cell(GO:1903410) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.4 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 1.2 | 3.5 | GO:0071754 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 1.0 | 4.0 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.8 | 2.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.5 | 1.5 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.4 | 2.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.4 | 4.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.4 | 2.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.3 | 1.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 1.0 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.3 | 3.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.3 | 13.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 5.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.3 | 1.6 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.3 | 1.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 2.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 1.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.2 | 1.8 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 16.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 1.0 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 1.6 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.2 | 1.8 | GO:0032133 | chromosome passenger complex(GO:0032133) meiotic spindle(GO:0072687) |
| 0.2 | 4.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 0.6 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.2 | 3.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 2.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 1.1 | GO:0005683 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.2 | 0.8 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 0.9 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.9 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 7.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.8 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 2.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 3.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 2.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.9 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 1.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 1.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.7 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 7.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.6 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 2.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.3 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 15.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 6.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.1 | 1.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 0.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.9 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.6 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 1.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 7.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 2.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.6 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.3 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 0.4 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.8 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 2.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.3 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 20.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 1.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 5.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.7 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 1.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 3.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 57.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.9 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 8.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.9 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 8.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 1.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 14.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 1.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.2 | 3.7 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 1.1 | 3.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.1 | 5.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.9 | 3.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.9 | 4.7 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.9 | 3.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.8 | 3.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.8 | 3.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.7 | 2.6 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.6 | 3.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.6 | 22.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.6 | 2.8 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.5 | 1.6 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.5 | 1.5 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.5 | 1.9 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.4 | 2.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.4 | 3.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.4 | 1.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.4 | 2.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.4 | 1.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.4 | 2.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.3 | 1.6 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.3 | 1.0 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.3 | 1.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 1.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 2.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.3 | 6.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 2.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 2.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.3 | 6.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.3 | 1.6 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.3 | 0.8 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.3 | 1.0 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.3 | 0.8 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.3 | 5.8 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 7.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.6 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 2.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 3.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.2 | 1.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 13.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 0.7 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.2 | 4.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 5.9 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 0.9 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 0.8 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.2 | 0.6 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.2 | 0.4 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 0.8 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.2 | 1.2 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.2 | 1.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 1.7 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) phosphate ion binding(GO:0042301) |
| 0.2 | 3.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 2.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 2.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 4.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 0.5 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.2 | 1.5 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 4.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 4.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 0.5 | GO:0003860 | 3-hydroxyisobutyryl-CoA hydrolase activity(GO:0003860) |
| 0.2 | 0.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 0.5 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.2 | 1.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 2.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 1.3 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 2.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.6 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 4.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.8 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.1 | 2.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.6 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 1.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 2.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 3.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.8 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 1.3 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.9 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.7 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.4 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 6.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 3.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 20.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 3.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.5 | GO:0038181 | bile acid receptor activity(GO:0038181) chenodeoxycholic acid binding(GO:1902122) |
| 0.1 | 0.5 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 1.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 1.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.6 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.4 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.1 | 1.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 1.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.3 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.1 | 1.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 2.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 2.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.5 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.5 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.7 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 1.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.3 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.1 | 0.3 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.4 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.9 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.4 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 5.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.7 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 10.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.6 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 1.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 5.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.2 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.1 | 0.2 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.1 | 0.9 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 6.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.8 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 4.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.6 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.2 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.8 | GO:1900750 | oligopeptide binding(GO:1900750) |
| 0.1 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 2.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.9 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 7.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.2 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.4 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 0.4 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.5 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 2.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 1.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 4.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.0 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 1.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 2.1 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.4 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 1.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 3.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 4.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 1.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 1.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 2.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.0 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.3 | GO:0008527 | taste receptor activity(GO:0008527) bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.6 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.6 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.7 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 6.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 2.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.0 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 2.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 5.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 0.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 2.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 4.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 5.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 3.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 7.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 5.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 5.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 16.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 4.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.1 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 5.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 2.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 9.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 5.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 21.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.6 | 7.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.5 | 7.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 6.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 7.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 3.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.3 | 4.0 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.3 | 5.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 2.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 2.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 3.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 2.8 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 3.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 5.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 3.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 3.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 3.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 2.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 2.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.2 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 5.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 12.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 6.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 3.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 3.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 3.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 8.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 6.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 3.9 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.4 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.2 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 2.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 2.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 2.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |