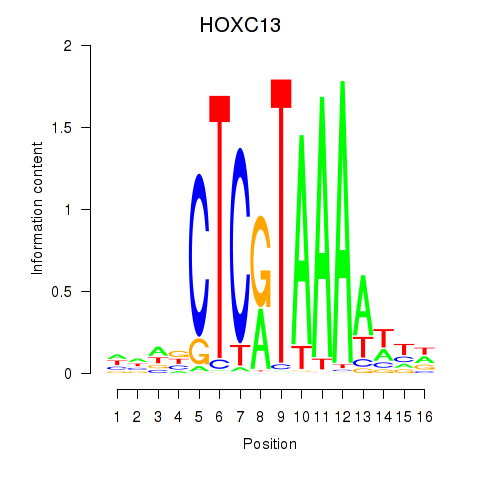
|
chr12_-_91146195
Show fit
|
40.55 |
ENST00000548218.1
|
DCN
|
decorin
|
|
chr3_-_195583931
Show fit
|
28.84 |
ENST00000343267.8
ENST00000421243.5
ENST00000453131.1
|
APOD
|
apolipoprotein D
|
|
chr4_+_105710809
Show fit
|
22.98 |
ENST00000360505.9
ENST00000510865.5
ENST00000509336.5
|
GSTCD
|
glutathione S-transferase C-terminal domain containing
|
|
chr2_-_86105839
Show fit
|
20.79 |
ENST00000263857.11
|
POLR1A
|
RNA polymerase I subunit A
|
|
chr5_-_79991237
Show fit
|
17.09 |
ENST00000512560.5
ENST00000509852.6
ENST00000512528.3
ENST00000617335.4
|
MTX3
|
metaxin 3
|
|
chr19_+_44905785
Show fit
|
14.09 |
ENST00000446996.5
ENST00000252486.9
ENST00000434152.5
|
APOE
|
apolipoprotein E
|
|
chr19_+_16888991
Show fit
|
13.39 |
ENST00000248076.4
|
F2RL3
|
F2R like thrombin or trypsin receptor 3
|
|
chr2_-_222656067
Show fit
|
12.78 |
ENST00000281828.8
|
FARSB
|
phenylalanyl-tRNA synthetase subunit beta
|
|
chr1_+_89995102
Show fit
|
12.50 |
ENST00000340281.9
ENST00000361911.9
ENST00000370447.3
|
ZNF326
|
zinc finger protein 326
|
|
chr12_-_16600703
Show fit
|
12.42 |
ENST00000616247.4
|
LMO3
|
LIM domain only 3
|
|
chr3_-_123620571
Show fit
|
12.29 |
ENST00000583087.5
|
MYLK
|
myosin light chain kinase
|
|
chr11_-_22829793
Show fit
|
11.89 |
ENST00000354193.5
|
SVIP
|
small VCP interacting protein
|
|
chr3_-_123620496
Show fit
|
11.51 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase
|
|
chr12_-_91153149
Show fit
|
11.26 |
ENST00000550758.1
|
DCN
|
decorin
|
|
chr3_+_98147479
Show fit
|
11.25 |
ENST00000641380.1
|
OR5H14
|
olfactory receptor family 5 subfamily H member 14
|
|
chr6_+_150721073
Show fit
|
10.61 |
ENST00000358517.6
|
PLEKHG1
|
pleckstrin homology and RhoGEF domain containing G1
|
|
chr3_-_179451387
Show fit
|
9.90 |
ENST00000675901.1
ENST00000232564.8
ENST00000674862.1
ENST00000497513.1
|
GNB4
|
G protein subunit beta 4
|
|
chr6_-_11807045
Show fit
|
9.49 |
ENST00000379415.6
|
ADTRP
|
androgen dependent TFPI regulating protein
|
|
chr19_-_9107475
Show fit
|
9.20 |
ENST00000641081.1
|
OR7G2
|
olfactory receptor family 7 subfamily G member 2
|
|
chr10_-_67838019
Show fit
|
8.99 |
ENST00000483798.6
|
DNAJC12
|
DnaJ heat shock protein family (Hsp40) member C12
|
|
chr15_+_70892443
Show fit
|
8.99 |
ENST00000443425.6
ENST00000560369.5
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr6_-_32371912
Show fit
|
8.83 |
ENST00000612031.4
|
TSBP1
|
testis expressed basic protein 1
|
|
chr11_+_62123991
Show fit
|
8.75 |
ENST00000533896.5
ENST00000278849.4
ENST00000394818.8
|
INCENP
|
inner centromere protein
|
|
chr20_-_54070520
Show fit
|
8.59 |
ENST00000371435.6
ENST00000395961.7
|
BCAS1
|
brain enriched myelin associated protein 1
|
|
chr6_-_32371897
Show fit
|
8.48 |
ENST00000442822.6
ENST00000375015.8
ENST00000533191.5
|
TSBP1
|
testis expressed basic protein 1
|
|
chr6_-_32371872
Show fit
|
8.45 |
ENST00000527965.5
ENST00000532023.5
ENST00000447241.6
ENST00000534588.1
|
TSBP1
|
testis expressed basic protein 1
|
|
chr11_-_5154757
Show fit
|
8.22 |
ENST00000380367.3
|
OR52A1
|
olfactory receptor family 52 subfamily A member 1
|
|
chr5_+_36596583
Show fit
|
8.06 |
ENST00000680318.1
|
SLC1A3
|
solute carrier family 1 member 3
|
|
chr6_-_52803807
Show fit
|
8.01 |
ENST00000334575.6
|
GSTA1
|
glutathione S-transferase alpha 1
|
|
chr6_-_52763473
Show fit
|
7.51 |
ENST00000493422.3
|
GSTA2
|
glutathione S-transferase alpha 2
|
|
chr4_+_94757921
Show fit
|
7.40 |
ENST00000515059.6
|
BMPR1B
|
bone morphogenetic protein receptor type 1B
|
|
chr6_-_52840843
Show fit
|
7.39 |
ENST00000370989.6
|
GSTA5
|
glutathione S-transferase alpha 5
|
|
chr4_-_1208825
Show fit
|
7.15 |
ENST00000511679.5
ENST00000617421.4
|
SPON2
|
spondin 2
|
|
chr9_+_98943705
Show fit
|
6.97 |
ENST00000610452.1
|
COL15A1
|
collagen type XV alpha 1 chain
|
|
chr6_-_49713521
Show fit
|
6.86 |
ENST00000339139.5
|
CRISP2
|
cysteine rich secretory protein 2
|
|
chr6_-_33271835
Show fit
|
6.79 |
ENST00000482399.5
ENST00000445902.3
|
VPS52
|
VPS52 subunit of GARP complex
|
|
chr2_+_102418642
Show fit
|
6.71 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr1_+_92080305
Show fit
|
6.62 |
ENST00000342818.4
ENST00000636805.2
|
BTBD8
|
BTB domain containing 8
|
|
chr15_+_70892809
Show fit
|
6.62 |
ENST00000260382.10
ENST00000560755.5
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr12_-_114403898
Show fit
|
6.57 |
ENST00000526441.1
|
TBX5
|
T-box transcription factor 5
|
|
chr6_-_49713564
Show fit
|
6.28 |
ENST00000616725.4
ENST00000618917.4
|
CRISP2
|
cysteine rich secretory protein 2
|
|
chr17_-_2511875
Show fit
|
6.27 |
ENST00000263092.11
ENST00000576976.2
|
METTL16
|
methyltransferase like 16
|
|
chr7_-_57132425
Show fit
|
6.24 |
ENST00000319636.10
|
ZNF479
|
zinc finger protein 479
|
|
chr15_-_70892412
Show fit
|
5.69 |
ENST00000249861.9
|
THAP10
|
THAP domain containing 10
|
|
chr5_+_36608146
Show fit
|
5.68 |
ENST00000381918.4
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 member 3
|
|
chr8_-_109680812
Show fit
|
5.63 |
ENST00000528716.5
ENST00000527600.5
ENST00000531230.5
ENST00000532189.5
ENST00000534184.5
ENST00000408889.7
ENST00000533171.5
|
SYBU
|
syntabulin
|
|
chr3_-_51499950
Show fit
|
5.50 |
ENST00000423656.5
ENST00000504652.5
ENST00000684031.1
|
DCAF1
|
DDB1 and CUL4 associated factor 1
|
|
chr10_-_77637789
Show fit
|
5.46 |
ENST00000481070.1
ENST00000640969.1
ENST00000286628.14
ENST00000638991.1
ENST00000639913.1
ENST00000480683.2
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr22_-_18936142
Show fit
|
5.45 |
ENST00000438924.5
ENST00000457083.1
ENST00000357068.11
ENST00000420436.5
ENST00000334029.6
ENST00000610940.4
|
PRODH
|
proline dehydrogenase 1
|
|
chr19_-_18522051
Show fit
|
5.40 |
ENST00000262809.9
|
ELL
|
elongation factor for RNA polymerase II
|
|
chr11_+_123902167
Show fit
|
5.35 |
ENST00000641687.1
|
OR8D4
|
olfactory receptor family 8 subfamily D member 4
|
|
chr1_-_46941425
Show fit
|
5.13 |
ENST00000371904.8
|
CYP4A11
|
cytochrome P450 family 4 subfamily A member 11
|
|
chr17_-_58692032
Show fit
|
5.13 |
ENST00000349033.10
ENST00000389934.7
|
TEX14
|
testis expressed 14, intercellular bridge forming factor
|
|
chr6_-_145735964
Show fit
|
4.94 |
ENST00000640980.1
ENST00000639423.1
ENST00000611340.5
|
EPM2A
|
EPM2A glucan phosphatase, laforin
|
|
chr10_-_77637902
Show fit
|
4.91 |
ENST00000286627.10
ENST00000639486.1
ENST00000640523.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chrX_+_41724174
Show fit
|
4.85 |
ENST00000302548.5
|
GPR82
|
G protein-coupled receptor 82
|
|
chr17_-_58692021
Show fit
|
4.79 |
ENST00000240361.12
|
TEX14
|
testis expressed 14, intercellular bridge forming factor
|
|
chr4_+_20251896
Show fit
|
4.74 |
ENST00000504154.6
|
SLIT2
|
slit guidance ligand 2
|
|
chr8_+_144798913
Show fit
|
4.70 |
ENST00000359971.4
ENST00000528012.5
|
ZNF517
|
zinc finger protein 517
|
|
chr1_-_149861210
Show fit
|
4.69 |
ENST00000579512.2
|
H4C15
|
H4 clustered histone 15
|
|
chr19_+_21082190
Show fit
|
4.67 |
ENST00000618422.1
ENST00000618008.4
ENST00000425625.5
ENST00000456283.7
|
ZNF714
|
zinc finger protein 714
|
|
chr12_-_9208388
Show fit
|
4.49 |
ENST00000261336.7
|
PZP
|
PZP alpha-2-macroglobulin like
|
|
chr3_-_98523013
Show fit
|
4.46 |
ENST00000394181.6
ENST00000508902.5
ENST00000394180.6
|
CLDND1
|
claudin domain containing 1
|
|
chr19_-_11346259
Show fit
|
4.46 |
ENST00000590788.1
ENST00000354882.10
ENST00000586590.5
ENST00000589555.5
ENST00000586956.5
ENST00000593256.6
ENST00000447337.5
ENST00000591677.5
ENST00000586701.1
ENST00000589655.1
|
TMEM205
RAB3D
|
transmembrane protein 205
RAB3D, member RAS oncogene family
|
|
chr5_-_65624288
Show fit
|
4.45 |
ENST00000381018.7
ENST00000274327.11
ENST00000231524.14
|
TRIM23
|
tripartite motif containing 23
|
|
chr16_+_81314922
Show fit
|
4.38 |
ENST00000648994.2
|
GAN
|
gigaxonin
|
|
chr7_+_134866831
Show fit
|
4.31 |
ENST00000435928.1
|
CALD1
|
caldesmon 1
|
|
chr9_-_5304713
Show fit
|
4.13 |
ENST00000381627.4
|
RLN2
|
relaxin 2
|
|
chr1_+_43300971
Show fit
|
3.95 |
ENST00000372476.8
ENST00000538015.1
|
TIE1
|
tyrosine kinase with immunoglobulin like and EGF like domains 1
|
|
chr18_-_5544250
Show fit
|
3.93 |
ENST00000540638.6
ENST00000342933.7
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3
|
|
chrX_+_49922559
Show fit
|
3.90 |
ENST00000376091.8
|
CLCN5
|
chloride voltage-gated channel 5
|
|
chr20_+_38317627
Show fit
|
3.89 |
ENST00000417318.3
|
BPI
|
bactericidal permeability increasing protein
|
|
chr11_+_101914997
Show fit
|
3.82 |
ENST00000263468.13
|
CEP126
|
centrosomal protein 126
|
|
chr10_-_46046264
Show fit
|
3.78 |
ENST00000581478.5
ENST00000582163.3
|
MSMB
|
microseminoprotein beta
|
|
chr5_-_161546671
Show fit
|
3.77 |
ENST00000517547.5
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2
|
|
chr12_-_52434363
Show fit
|
3.75 |
ENST00000252245.6
|
KRT75
|
keratin 75
|
|
chr10_-_68527498
Show fit
|
3.68 |
ENST00000609923.6
|
SLC25A16
|
solute carrier family 25 member 16
|
|
chr8_+_132866948
Show fit
|
3.65 |
ENST00000220616.9
|
TG
|
thyroglobulin
|
|
chr1_-_46941464
Show fit
|
3.61 |
ENST00000462347.5
ENST00000371905.1
ENST00000310638.9
|
CYP4A11
|
cytochrome P450 family 4 subfamily A member 11
|
|
chr3_-_99850976
Show fit
|
3.61 |
ENST00000487087.5
|
FILIP1L
|
filamin A interacting protein 1 like
|
|
chr22_+_39520553
Show fit
|
3.55 |
ENST00000674920.3
ENST00000679776.1
ENST00000675582.2
ENST00000337304.2
ENST00000676346.2
ENST00000396680.3
ENST00000680446.1
ENST00000674568.2
ENST00000680748.1
ENST00000674835.2
|
ATF4
|
activating transcription factor 4
|
|
chr15_-_78245683
Show fit
|
3.53 |
ENST00000560817.5
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1
|
|
chr12_-_118359105
Show fit
|
3.50 |
ENST00000541186.5
ENST00000539872.5
|
TAOK3
|
TAO kinase 3
|
|
chr12_+_64780465
Show fit
|
3.49 |
ENST00000542120.6
|
TBC1D30
|
TBC1 domain family member 30
|
|
chr11_+_66510626
Show fit
|
3.44 |
ENST00000526815.5
ENST00000318312.12
ENST00000525809.5
ENST00000455748.6
ENST00000393994.4
ENST00000630659.2
|
BBS1
|
Bardet-Biedl syndrome 1
|
|
chr4_+_87799546
Show fit
|
3.41 |
ENST00000226284.7
|
IBSP
|
integrin binding sialoprotein
|
|
chr11_-_124800630
Show fit
|
3.40 |
ENST00000239614.8
ENST00000674284.1
|
MSANTD2
|
Myb/SANT DNA binding domain containing 2
|
|
chr11_+_63838902
Show fit
|
3.38 |
ENST00000377810.8
|
MARK2
|
microtubule affinity regulating kinase 2
|
|
chr1_+_149832647
Show fit
|
3.37 |
ENST00000578186.2
|
H4C14
|
H4 clustered histone 14
|
|
chr2_+_209579399
Show fit
|
3.33 |
ENST00000360351.8
|
MAP2
|
microtubule associated protein 2
|
|
chr5_+_150640652
Show fit
|
3.32 |
ENST00000307662.5
|
SYNPO
|
synaptopodin
|
|
chr11_+_49028823
Show fit
|
3.31 |
ENST00000332682.9
|
TRIM49B
|
tripartite motif containing 49B
|
|
chr4_+_71062642
Show fit
|
3.26 |
ENST00000649996.1
|
SLC4A4
|
solute carrier family 4 member 4
|
|
chr17_-_41397600
Show fit
|
3.20 |
ENST00000251645.3
|
KRT31
|
keratin 31
|
|
chr19_-_23687163
Show fit
|
3.20 |
ENST00000601010.5
ENST00000601935.5
ENST00000600313.5
ENST00000596211.5
ENST00000359788.9
ENST00000599168.1
|
ZNF675
|
zinc finger protein 675
|
|
chr6_-_46954922
Show fit
|
3.17 |
ENST00000265417.7
|
ADGRF5
|
adhesion G protein-coupled receptor F5
|
|
chr15_-_58279245
Show fit
|
3.14 |
ENST00000558231.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2
|
|
chr7_+_106865263
Show fit
|
3.12 |
ENST00000440650.6
ENST00000496166.6
ENST00000473541.5
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma
|
|
chr4_+_41613476
Show fit
|
3.10 |
ENST00000508466.1
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr12_+_18262730
Show fit
|
3.08 |
ENST00000675017.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma
|
|
chr4_+_69995958
Show fit
|
3.06 |
ENST00000381060.2
ENST00000246895.9
|
STATH
|
statherin
|
|
chr8_-_94262308
Show fit
|
3.02 |
ENST00000297596.3
ENST00000396194.6
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr8_-_17697654
Show fit
|
3.02 |
ENST00000297488.10
|
MTUS1
|
microtubule associated scaffold protein 1
|
|
chr1_+_54641806
Show fit
|
3.01 |
ENST00000409996.5
|
MROH7
|
maestro heat like repeat family member 7
|
|
chr2_+_161160299
Show fit
|
3.00 |
ENST00000440506.5
ENST00000429217.5
ENST00000406287.5
ENST00000402568.5
|
TANK
|
TRAF family member associated NFKB activator
|
|
chr1_+_54641754
Show fit
|
2.97 |
ENST00000339553.9
ENST00000421030.7
|
MROH7
|
maestro heat like repeat family member 7
|
|
chr15_-_99249523
Show fit
|
2.96 |
ENST00000560235.1
ENST00000394132.7
ENST00000560860.5
ENST00000558078.5
ENST00000560772.5
|
TTC23
|
tetratricopeptide repeat domain 23
|
|
chr12_-_13095628
Show fit
|
2.91 |
ENST00000457134.6
ENST00000537302.5
|
GSG1
|
germ cell associated 1
|
|
chr1_+_153031195
Show fit
|
2.91 |
ENST00000307098.5
|
SPRR1B
|
small proline rich protein 1B
|
|
chr3_+_129440196
Show fit
|
2.90 |
ENST00000507564.5
ENST00000348417.7
ENST00000504021.5
ENST00000349441.6
|
IFT122
|
intraflagellar transport 122
|
|
chr12_-_52814106
Show fit
|
2.89 |
ENST00000551956.2
|
KRT4
|
keratin 4
|
|
chr12_-_13095664
Show fit
|
2.81 |
ENST00000337630.10
ENST00000545699.1
|
GSG1
|
germ cell associated 1
|
|
chr4_-_69961007
Show fit
|
2.78 |
ENST00000353151.3
|
CSN2
|
casein beta
|
|
chr19_-_6279921
Show fit
|
2.77 |
ENST00000252674.9
|
MLLT1
|
MLLT1 super elongation complex subunit
|
|
chr5_-_180649579
Show fit
|
2.73 |
ENST00000261937.11
ENST00000502649.5
|
FLT4
|
fms related receptor tyrosine kinase 4
|
|
chr5_-_161546708
Show fit
|
2.66 |
ENST00000393959.6
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2
|
|
chr2_+_137964446
Show fit
|
2.63 |
ENST00000280096.5
ENST00000280097.5
|
HNMT
|
histamine N-methyltransferase
|
|
chr7_-_38349972
Show fit
|
2.57 |
ENST00000390344.2
|
TRGV5
|
T cell receptor gamma variable 5
|
|
chr3_-_126655046
Show fit
|
2.56 |
ENST00000523403.3
ENST00000524230.9
|
TXNRD3
|
thioredoxin reductase 3
|
|
chr11_+_89924064
Show fit
|
2.55 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2
|
|
chr2_+_209579429
Show fit
|
2.53 |
ENST00000361559.8
|
MAP2
|
microtubule associated protein 2
|
|
chr2_+_209579598
Show fit
|
2.48 |
ENST00000445941.5
ENST00000673860.1
|
MAP2
|
microtubule associated protein 2
|
|
chr3_+_159839847
Show fit
|
2.47 |
ENST00000445224.6
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr5_-_161546970
Show fit
|
2.45 |
ENST00000675303.1
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2
|
|
chr1_-_46176482
Show fit
|
2.43 |
ENST00000540385.2
ENST00000506599.2
|
P3R3URF-PIK3R3
P3R3URF
|
P3R3URF-PIK3R3 readthrough
PIK3R3 upstream reading frame
|
|
chr11_+_61248122
Show fit
|
2.39 |
ENST00000451616.6
|
PGA5
|
pepsinogen A5
|
|
chr1_+_50105666
Show fit
|
2.38 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4
|
|
chr11_+_30231000
Show fit
|
2.34 |
ENST00000254122.8
ENST00000533718.2
ENST00000417547.1
|
FSHB
|
follicle stimulating hormone subunit beta
|
|
chr11_+_62270150
Show fit
|
2.29 |
ENST00000525380.1
ENST00000227918.3
|
SCGB2A2
|
secretoglobin family 2A member 2
|
|
chr5_+_140868945
Show fit
|
2.29 |
ENST00000398640.7
|
PCDHA11
|
protocadherin alpha 11
|
|
chr19_+_21082140
Show fit
|
2.25 |
ENST00000616183.1
ENST00000596053.5
|
ZNF714
|
zinc finger protein 714
|
|
chrX_+_114584037
Show fit
|
2.25 |
ENST00000371951.5
ENST00000371950.3
ENST00000276198.6
|
HTR2C
|
5-hydroxytryptamine receptor 2C
|
|
chr12_+_130162456
Show fit
|
2.24 |
ENST00000539839.1
ENST00000229030.5
|
FZD10
|
frizzled class receptor 10
|
|
chr1_+_47137435
Show fit
|
2.24 |
ENST00000371891.8
ENST00000371890.7
ENST00000619754.4
ENST00000294337.7
ENST00000620131.1
|
CYP4A22
|
cytochrome P450 family 4 subfamily A member 22
|
|
chr18_+_63887698
Show fit
|
2.23 |
ENST00000457692.5
ENST00000299502.9
ENST00000413956.5
|
SERPINB2
|
serpin family B member 2
|
|
chr22_+_22758698
Show fit
|
2.21 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14
|
|
chr12_-_4538440
Show fit
|
2.21 |
ENST00000261250.8
ENST00000541014.5
ENST00000545746.5
ENST00000542080.5
|
C12orf4
|
chromosome 12 open reading frame 4
|
|
chr4_+_153152163
Show fit
|
2.19 |
ENST00000676423.1
ENST00000675745.1
ENST00000676348.1
ENST00000676408.1
ENST00000674874.1
ENST00000675315.1
ENST00000675518.1
|
TRIM2
ENSG00000288637.1
|
tripartite motif containing 2
novel protein
|
|
chr15_+_70936487
Show fit
|
2.18 |
ENST00000558456.5
ENST00000560158.6
ENST00000558808.5
ENST00000559806.5
ENST00000559069.1
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr7_+_106865474
Show fit
|
2.15 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma
|
|
chr9_-_101384999
Show fit
|
2.15 |
ENST00000259407.7
|
BAAT
|
bile acid-CoA:amino acid N-acyltransferase
|
|
chr1_-_54801290
Show fit
|
2.14 |
ENST00000371276.9
|
TTC22
|
tetratricopeptide repeat domain 22
|
|
chr22_-_19447686
Show fit
|
2.12 |
ENST00000399568.5
ENST00000399562.9
|
C22orf39
|
chromosome 22 open reading frame 39
|
|
chr7_-_16833411
Show fit
|
2.10 |
ENST00000412973.1
|
AGR2
|
anterior gradient 2, protein disulphide isomerase family member
|
|
chr5_+_102808057
Show fit
|
2.10 |
ENST00000684043.1
ENST00000682407.1
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr4_+_89901979
Show fit
|
2.07 |
ENST00000508372.1
|
MMRN1
|
multimerin 1
|
|
chr19_+_20923222
Show fit
|
2.01 |
ENST00000597314.5
ENST00000328178.13
ENST00000601924.5
|
ZNF85
|
zinc finger protein 85
|
|
chr5_+_141402764
Show fit
|
2.01 |
ENST00000573521.2
ENST00000616887.1
|
PCDHGA9
|
protocadherin gamma subfamily A, 9
|
|
chr1_+_213051229
Show fit
|
1.98 |
ENST00000615329.4
ENST00000543354.5
ENST00000614059.4
ENST00000543470.5
ENST00000366960.8
ENST00000366959.4
|
RPS6KC1
|
ribosomal protein S6 kinase C1
|
|
chr3_+_155083889
Show fit
|
1.95 |
ENST00000680282.1
|
MME
|
membrane metalloendopeptidase
|
|
chr22_+_18150162
Show fit
|
1.90 |
ENST00000215794.8
|
USP18
|
ubiquitin specific peptidase 18
|
|
chr11_+_61228377
Show fit
|
1.83 |
ENST00000537932.5
|
PGA4
|
pepsinogen A4
|
|
chr10_-_77637721
Show fit
|
1.81 |
ENST00000638848.1
ENST00000639406.1
ENST00000618048.2
ENST00000639120.1
ENST00000640834.1
ENST00000639601.1
ENST00000638514.1
ENST00000457953.6
ENST00000639090.1
ENST00000639489.1
ENST00000372440.6
ENST00000404771.8
ENST00000638203.1
ENST00000638306.1
ENST00000638351.1
ENST00000638606.1
ENST00000639591.1
ENST00000640182.1
ENST00000640605.1
ENST00000640141.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr9_+_101185029
Show fit
|
1.77 |
ENST00000395056.2
|
PLPPR1
|
phospholipid phosphatase related 1
|
|
chr3_+_155083523
Show fit
|
1.76 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase
|
|
chr3_+_148791058
Show fit
|
1.75 |
ENST00000491148.5
|
CPB1
|
carboxypeptidase B1
|
|
chr10_-_77638369
Show fit
|
1.71 |
ENST00000372443.6
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr1_+_67307360
Show fit
|
1.70 |
ENST00000262345.5
ENST00000544434.5
|
IL12RB2
|
interleukin 12 receptor subunit beta 2
|
|
chr20_+_38962299
Show fit
|
1.69 |
ENST00000373325.6
ENST00000373323.8
ENST00000252011.8
ENST00000615559.1
|
DHX35
ENSG00000277581.1
|
DEAH-box helicase 35
novel transcript, sense intronic to DHX35
|
|
chr16_-_72172135
Show fit
|
1.65 |
ENST00000537465.5
ENST00000237353.15
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr12_-_57237090
Show fit
|
1.64 |
ENST00000556732.1
|
NDUFA4L2
|
NDUFA4 mitochondrial complex associated like 2
|
|
chr5_-_180649613
Show fit
|
1.63 |
ENST00000393347.7
ENST00000619105.4
|
FLT4
|
fms related receptor tyrosine kinase 4
|
|
chr3_-_52830664
Show fit
|
1.62 |
ENST00000266041.9
ENST00000406595.5
ENST00000485816.5
|
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain 4
|
|
chr9_-_32526185
Show fit
|
1.58 |
ENST00000379883.3
ENST00000379868.6
ENST00000679859.1
|
DDX58
|
DExD/H-box helicase 58
|
|
chr4_-_68670648
Show fit
|
1.56 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15
|
|
chr17_-_3592877
Show fit
|
1.52 |
ENST00000399756.8
|
TRPV1
|
transient receptor potential cation channel subfamily V member 1
|
|
chr11_+_22668101
Show fit
|
1.43 |
ENST00000630668.2
ENST00000278187.7
|
GAS2
|
growth arrest specific 2
|
|
chr5_-_22853320
Show fit
|
1.43 |
ENST00000504376.6
ENST00000382254.6
|
CDH12
|
cadherin 12
|
|
chr1_-_89126066
Show fit
|
1.42 |
ENST00000370466.4
|
GBP2
|
guanylate binding protein 2
|
|
chr14_+_96256194
Show fit
|
1.42 |
ENST00000216629.11
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1
|
|
chr12_-_55296569
Show fit
|
1.39 |
ENST00000358433.3
|
OR6C6
|
olfactory receptor family 6 subfamily C member 6
|
|
chr19_+_11239602
Show fit
|
1.39 |
ENST00000252453.12
|
ANGPTL8
|
angiopoietin like 8
|
|
chr4_-_119322128
Show fit
|
1.35 |
ENST00000274024.4
|
FABP2
|
fatty acid binding protein 2
|
|
chr5_-_59276109
Show fit
|
1.25 |
ENST00000503258.5
|
PDE4D
|
phosphodiesterase 4D
|
|
chr9_+_114611206
Show fit
|
1.25 |
ENST00000374049.4
ENST00000288502.9
|
TMEM268
|
transmembrane protein 268
|
|
chr8_-_85378105
Show fit
|
1.14 |
ENST00000521846.5
ENST00000523022.6
ENST00000524324.5
ENST00000519991.5
ENST00000520663.5
ENST00000517590.5
ENST00000522579.5
ENST00000522814.5
ENST00000522662.5
ENST00000523858.5
ENST00000519129.5
|
CA1
|
carbonic anhydrase 1
|
|
chr10_-_67696115
Show fit
|
1.12 |
ENST00000433211.7
|
CTNNA3
|
catenin alpha 3
|
|
chr10_-_20897288
Show fit
|
1.10 |
ENST00000377122.9
|
NEBL
|
nebulette
|
|
chr17_-_41382298
Show fit
|
1.03 |
ENST00000394001.3
|
KRT34
|
keratin 34
|
|
chr1_+_50106265
Show fit
|
1.02 |
ENST00000357083.8
|
ELAVL4
|
ELAV like RNA binding protein 4
|
|
chr16_-_20352707
Show fit
|
0.97 |
ENST00000396134.6
ENST00000573567.5
ENST00000570757.5
ENST00000396138.9
ENST00000571174.5
ENST00000576688.2
|
UMOD
|
uromodulin
|
|
chr3_+_190615308
Show fit
|
0.90 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein
|
|
chr2_+_102337148
Show fit
|
0.85 |
ENST00000311734.6
ENST00000409584.5
|
IL1RL1
|
interleukin 1 receptor like 1
|
|
chrX_+_47370578
Show fit
|
0.84 |
ENST00000377073.4
|
ZNF157
|
zinc finger protein 157
|
|
chr3_+_178419123
Show fit
|
0.83 |
ENST00000614557.1
ENST00000455307.5
ENST00000436432.1
|
ENSG00000275163.1
LINC01014
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 (KCNMB2-IT1 - KCNMB2 readthrough transcript)
long intergenic non-protein coding RNA 1014
|
|
chr19_+_21142024
Show fit
|
0.78 |
ENST00000600692.5
ENST00000599296.5
ENST00000594425.5
ENST00000311048.11
|
ZNF431
|
zinc finger protein 431
|
|
chr21_+_38256698
Show fit
|
0.74 |
ENST00000613499.4
ENST00000612702.4
ENST00000398925.5
ENST00000398928.5
ENST00000328656.8
ENST00000443341.5
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15
|
|
chr5_+_42548043
Show fit
|
0.74 |
ENST00000618088.4
ENST00000612382.4
|
GHR
|
growth hormone receptor
|
|
chr19_-_37594746
Show fit
|
0.73 |
ENST00000593133.5
ENST00000590751.5
ENST00000358744.3
ENST00000328550.6
ENST00000451802.7
|
ZNF571
|
zinc finger protein 571
|
|
chr7_+_57450171
Show fit
|
0.72 |
ENST00000420713.2
|
ZNF716
|
zinc finger protein 716
|
|
chr6_-_117425905
Show fit
|
0.67 |
ENST00000368507.8
|
ROS1
|
ROS proto-oncogene 1, receptor tyrosine kinase
|
|
chr5_-_39270623
Show fit
|
0.61 |
ENST00000512138.1
ENST00000646045.2
|
FYB1
|
FYN binding protein 1
|
|
chr19_+_13747887
Show fit
|
0.61 |
ENST00000221554.13
ENST00000586666.5
|
CCDC130
|
coiled-coil domain containing 130
|
|
chr1_-_154178174
Show fit
|
0.60 |
ENST00000302206.9
|
TPM3
|
tropomyosin 3
|
|
chr9_-_92482350
Show fit
|
0.58 |
ENST00000375543.2
|
ASPN
|
asporin
|
|
chr4_+_68815991
Show fit
|
0.57 |
ENST00000265403.12
ENST00000458688.2
|
UGT2B10
|
UDP glucuronosyltransferase family 2 member B10
|
|
chrX_-_100874332
Show fit
|
0.48 |
ENST00000372960.8
|
NOX1
|
NADPH oxidase 1
|
|
chr4_+_69280472
Show fit
|
0.43 |
ENST00000335568.10
ENST00000511240.1
|
UGT2B28
|
UDP glucuronosyltransferase family 2 member B28
|
|
chr17_-_10469558
Show fit
|
0.41 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4
|
|
chr6_+_20534441
Show fit
|
0.38 |
ENST00000274695.8
ENST00000613575.4
|
CDKAL1
|
CDK5 regulatory subunit associated protein 1 like 1
|