Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
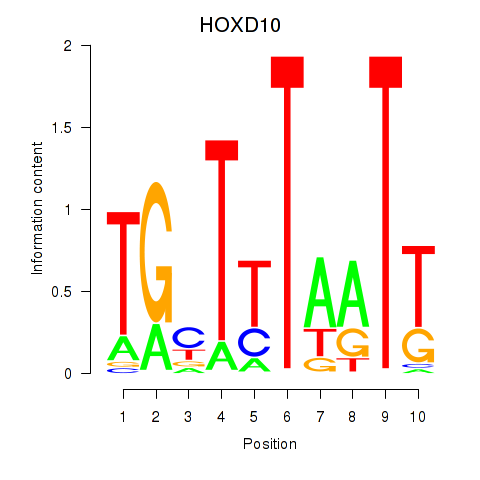
Results for HOXD10
Z-value: 0.43
Transcription factors associated with HOXD10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD10
|
ENSG00000128710.6 | HOXD10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD10 | hg38_v1_chr2_+_176116768_176116794 | -0.19 | 5.8e-03 | Click! |
Activity profile of HOXD10 motif
Sorted Z-values of HOXD10 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 13.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 3.8 | 15.2 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 2.3 | 29.6 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 1.6 | 4.9 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 1.6 | 4.9 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 1.4 | 5.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 1.2 | 3.6 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 1.1 | 4.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.1 | 6.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 1.0 | 5.1 | GO:2001107 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 1.0 | 7.8 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.9 | 12.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.8 | 10.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.8 | 2.3 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.7 | 5.8 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.7 | 4.3 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.7 | 4.7 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.6 | 2.6 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.6 | 6.3 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.6 | 11.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.6 | 2.4 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.6 | 2.3 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.5 | 2.0 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.5 | 2.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.5 | 1.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.4 | 1.3 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.4 | 1.2 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.4 | 1.6 | GO:0061643 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.4 | 2.3 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.4 | 4.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.4 | 2.2 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.4 | 1.5 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.4 | 4.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.3 | 1.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 1.7 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.3 | 1.3 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.3 | 1.9 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.3 | 1.8 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.3 | 3.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.3 | 8.5 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.3 | 7.7 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.3 | 1.9 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 1.9 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.3 | 1.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.3 | 3.4 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.3 | 3.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 1.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.3 | 1.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 16.5 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.2 | 2.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 2.0 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.2 | 1.8 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.2 | 0.7 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.2 | 0.2 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 2.0 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 1.3 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.2 | 3.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 1.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 1.0 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.2 | 2.1 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.2 | 0.9 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 | 1.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 8.8 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.2 | 1.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 1.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 1.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 1.0 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.2 | 1.0 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.2 | 0.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 1.7 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 17.1 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 3.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 2.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 3.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 1.7 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 3.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 2.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.1 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.1 | 0.4 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.1 | 0.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 2.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.4 | GO:0022018 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.1 | 2.0 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.5 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 0.7 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 18.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.0 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 4.5 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 2.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 1.3 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 1.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.9 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 1.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 7.7 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 10.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.9 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.1 | 4.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.3 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 3.8 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 5.3 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 3.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.9 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.4 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.3 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 5.2 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.1 | 0.4 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 5.5 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 0.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 1.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.2 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.1 | 2.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 2.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.9 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 1.2 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 6.8 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 0.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 1.1 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 2.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 0.4 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.8 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 3.4 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.6 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.7 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 2.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.0 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 1.0 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0090238 | regulation of arachidonic acid secretion(GO:0090237) positive regulation of arachidonic acid secretion(GO:0090238) regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.8 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 0.5 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 3.4 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 1.4 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.1 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.0 | 2.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 1.4 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 1.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 5.0 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 1.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.9 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 2.7 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.7 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 0.0 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.0 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.6 | GO:0006400 | tRNA modification(GO:0006400) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.7 | 32.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.5 | 4.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.2 | 29.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 1.0 | 4.9 | GO:0031523 | Myb complex(GO:0031523) |
| 1.0 | 5.8 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.8 | 3.4 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.8 | 4.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.8 | 10.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.6 | 1.9 | GO:0097414 | glial cytoplasmic inclusion(GO:0097409) classical Lewy body(GO:0097414) Lewy neurite(GO:0097462) Lewy body corona(GO:1990038) |
| 0.6 | 4.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.6 | 1.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 2.2 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.5 | 4.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.5 | 1.5 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.5 | 6.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.5 | 5.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.4 | 5.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.4 | 1.3 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.3 | 1.3 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.3 | 2.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 3.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.0 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 3.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 3.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 1.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 13.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 1.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 8.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 2.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 2.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 2.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 5.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 0.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 10.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.4 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 6.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 11.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 0.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 1.0 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.9 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 12.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 5.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 25.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.2 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.7 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 2.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 1.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 23.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.9 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 5.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 5.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 1.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0001940 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.0 | 1.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 2.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 2.6 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 4.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.8 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 4.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.3 | GO:0005903 | brush border(GO:0005903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 1.3 | 29.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.1 | 4.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 1.0 | 3.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 1.0 | 3.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 1.0 | 4.9 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.8 | 32.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.8 | 2.4 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.7 | 5.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.7 | 16.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.6 | 1.9 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.6 | 4.7 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.5 | 3.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.4 | 1.8 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.4 | 2.3 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.4 | 2.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.4 | 1.9 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.3 | 10.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 2.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.3 | 1.9 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.3 | 1.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 1.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 5.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.3 | 11.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.3 | 1.6 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.3 | 1.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.3 | 0.8 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 1.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.2 | 6.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 1.6 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.2 | 5.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 2.6 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.2 | 7.0 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.2 | 3.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 2.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 8.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 2.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 2.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 3.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 2.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.4 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.1 | 5.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 2.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 10.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 2.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 0.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 2.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 1.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 3.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 3.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 3.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.9 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 16.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 6.6 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 0.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) glycosphingolipid binding(GO:0043208) |
| 0.1 | 3.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.8 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 1.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 4.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 2.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 1.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.7 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.1 | 3.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 2.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 2.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 2.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 5.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 1.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 1.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 3.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 2.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 2.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 5.1 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 1.0 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 2.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 8.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 3.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 14.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 3.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 6.7 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 5.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.7 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 4.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 4.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 39.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.3 | 3.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 12.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 11.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 4.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 8.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 5.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 5.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 3.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 1.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 6.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 39.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.8 | 29.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.4 | 4.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 10.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.3 | 4.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 3.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 2.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 4.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 3.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 5.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 3.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 1.5 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 6.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 4.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 3.8 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 1.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 19.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.6 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 6.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 3.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 3.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 4.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 3.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.0 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 4.4 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 1.5 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |