Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for HSF1
Z-value: 0.91
Transcription factors associated with HSF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
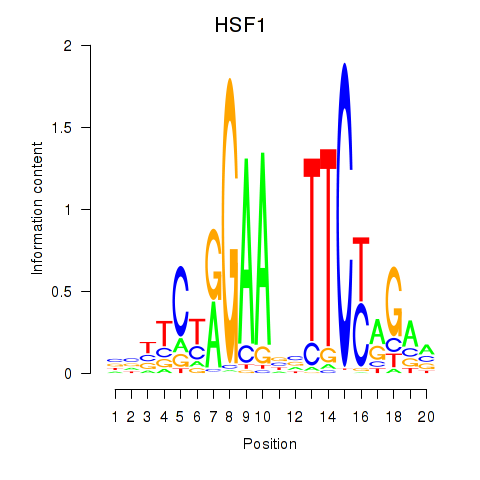
HSF1
|
ENSG00000185122.11 | HSF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF1 | hg38_v1_chr8_+_144291581_144291617 | 0.00 | 9.6e-01 | Click! |
Activity profile of HSF1 motif
Sorted Z-values of HSF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.3 | GO:0002368 | B cell cytokine production(GO:0002368) |
| 2.5 | 7.6 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 1.5 | 4.6 | GO:1903487 | positive regulation of vascular wound healing(GO:0035470) regulation of lactation(GO:1903487) |
| 1.5 | 6.0 | GO:1900190 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) positive regulation of chondrocyte proliferation(GO:1902732) |
| 1.5 | 7.4 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 1.4 | 4.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 1.3 | 3.9 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 1.2 | 3.6 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 1.0 | 10.4 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 1.0 | 5.8 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.9 | 2.8 | GO:0014810 | positive regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014810) |
| 0.9 | 3.6 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.9 | 4.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.9 | 2.6 | GO:0038156 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.8 | 2.5 | GO:1902232 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) regulation of positive thymic T cell selection(GO:1902232) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.8 | 2.5 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.8 | 3.1 | GO:0050705 | negative regulation of interleukin-1 alpha production(GO:0032690) regulation of interleukin-1 alpha secretion(GO:0050705) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.7 | 1.4 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.7 | 12.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.7 | 8.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.7 | 4.0 | GO:0010266 | response to vitamin B1(GO:0010266) |
| 0.7 | 2.0 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.7 | 2.0 | GO:2000547 | mesangial cell-matrix adhesion(GO:0035759) dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.7 | 2.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.6 | 1.2 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.6 | 3.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.6 | 14.9 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.6 | 3.5 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.6 | 4.0 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.6 | 7.2 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.5 | 3.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.5 | 2.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.5 | 5.9 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.5 | 1.6 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.5 | 3.6 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.5 | 2.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.5 | 2.0 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.5 | 1.0 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.5 | 1.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.5 | 1.9 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.5 | 1.4 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.5 | 0.9 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.5 | 2.8 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.5 | 5.4 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.4 | 2.6 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.4 | 0.9 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.4 | 6.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.4 | 6.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.4 | 11.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 1.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.4 | 11.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.4 | 1.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.4 | 1.5 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.4 | 5.2 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.4 | 8.0 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.4 | 1.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.4 | 2.5 | GO:0015866 | ADP transport(GO:0015866) |
| 0.3 | 2.8 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.3 | 1.7 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.3 | 2.7 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.3 | 5.0 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 | 1.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.3 | 1.3 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.3 | 3.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.3 | 3.0 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.3 | 0.3 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.3 | 0.9 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.3 | 2.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.3 | 0.9 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.3 | 7.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 1.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.3 | 7.8 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.3 | 1.7 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.3 | 3.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.3 | 2.7 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.3 | 1.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.3 | 3.7 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.3 | 1.3 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.3 | 1.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 2.0 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.2 | 1.9 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.2 | 1.5 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 2.9 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.2 | 1.7 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 0.7 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.2 | 0.5 | GO:0002415 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.2 | 1.4 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.2 | 1.3 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 0.7 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.2 | 0.6 | GO:0018963 | phthalate metabolic process(GO:0018963) epinephrine metabolic process(GO:0042414) epinephrine biosynthetic process(GO:0042418) phytoalexin metabolic process(GO:0052314) |
| 0.2 | 0.6 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.2 | 1.6 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.2 | 1.6 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.2 | 0.9 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 1.7 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.2 | 2.6 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.2 | 3.0 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.2 | 12.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 2.7 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.2 | 0.7 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.2 | 0.7 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 0.7 | GO:2000909 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.2 | 1.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 7.2 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.2 | 20.5 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.2 | 0.7 | GO:0002604 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.2 | 2.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.2 | 1.3 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 0.5 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.2 | 1.8 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.2 | 2.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 1.2 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.2 | 1.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 3.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 3.7 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 3.0 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.1 | 7.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 5.0 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.1 | 1.4 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 2.7 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.1 | 1.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.9 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 1.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 11.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.3 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.1 | 2.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.5 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.1 | 2.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 1.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.5 | GO:0009838 | abscission(GO:0009838) |
| 0.1 | 4.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.4 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 1.3 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 1.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 1.6 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 | 0.8 | GO:0051970 | negative regulation of action potential(GO:0045759) negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 1.5 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 2.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 1.3 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.1 | 0.2 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.1 | 2.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 9.0 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 0.9 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.7 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 2.5 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 2.6 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.1 | 0.2 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.1 | 6.6 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.1 | 2.7 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.1 | 1.6 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 4.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 2.4 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.6 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.1 | 0.6 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 3.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.0 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 7.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.6 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 1.6 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.4 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.9 | GO:0007140 | male meiosis(GO:0007140) |
| 0.1 | 24.7 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.7 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.2 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.1 | 1.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 5.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.1 | GO:0044146 | negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.1 | 4.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.4 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.1 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 2.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 4.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.1 | 3.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 2.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 4.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.7 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.5 | GO:0071492 | response to UV-A(GO:0070141) cellular response to UV-A(GO:0071492) |
| 0.0 | 1.9 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.0 | 1.5 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 1.0 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 2.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 1.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.9 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.2 | GO:0071110 | protein biotinylation(GO:0009305) histone biotinylation(GO:0071110) |
| 0.0 | 0.2 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.4 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 1.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 1.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.5 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.4 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.4 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 2.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.5 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.4 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.5 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 1.8 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 5.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 3.1 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.6 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.5 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.9 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 5.5 | GO:0007283 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0006308 | DNA catabolic process(GO:0006308) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 1.3 | 8.0 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 1.2 | 23.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.0 | 4.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.9 | 8.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.9 | 2.6 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.6 | 1.7 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.5 | 10.0 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.5 | 1.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.4 | 5.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.4 | 18.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 1.6 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.3 | 3.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 10.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.3 | 2.0 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.3 | 1.4 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.3 | 1.6 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.3 | 5.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 1.8 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 17.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 2.0 | GO:0032059 | bleb(GO:0032059) |
| 0.2 | 2.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 1.9 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 0.9 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 12.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 2.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 3.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 1.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 3.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 19.3 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 1.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 17.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 0.6 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.1 | 0.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.5 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 0.7 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 22.7 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 5.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.6 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.6 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 1.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.0 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 5.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.8 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 2.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 6.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 5.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 6.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 6.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.1 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 7.7 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.7 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 6.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.3 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 3.1 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 13.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 8.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 1.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 3.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 3.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 2.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 5.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.7 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 8.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 2.0 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 2.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 1.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 3.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 2.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 17.3 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 3.3 | 10.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 2.0 | 5.9 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 1.8 | 9.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.4 | 7.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 1.4 | 8.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 1.2 | 5.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 1.2 | 3.6 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 1.2 | 3.5 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 1.1 | 16.0 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 1.1 | 3.3 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 1.1 | 6.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.0 | 2.9 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.9 | 2.8 | GO:0047315 | kynurenine-glyoxylate transaminase activity(GO:0047315) |
| 0.9 | 2.6 | GO:0004914 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.9 | 1.7 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.8 | 2.5 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.6 | 3.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.6 | 1.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.6 | 4.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.6 | 3.0 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.6 | 4.0 | GO:0005497 | androgen binding(GO:0005497) |
| 0.6 | 2.8 | GO:0050610 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.5 | 2.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.5 | 1.6 | GO:0032428 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.5 | 1.6 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.5 | 3.6 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.5 | 4.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.5 | 2.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.5 | 8.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.5 | 3.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.5 | 2.3 | GO:0070915 | bioactive lipid receptor activity(GO:0045125) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.4 | 1.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.4 | 1.8 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.4 | 1.3 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.4 | 1.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.4 | 2.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.4 | 1.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.4 | 2.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.4 | 3.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.4 | 1.9 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.4 | 1.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.4 | 1.5 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.4 | 2.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.4 | 2.9 | GO:0019863 | IgE binding(GO:0019863) |
| 0.4 | 2.5 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 0.4 | 1.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.3 | 1.4 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.3 | 4.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 3.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 1.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.3 | 3.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.3 | 1.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.3 | 2.0 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.3 | 3.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.3 | 1.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 1.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 1.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 2.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 7.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 0.7 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 1.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 0.9 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.2 | 2.5 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.2 | 5.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 0.6 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.2 | 1.4 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.2 | 17.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 12.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 3.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 3.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 4.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 2.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 1.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 6.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 6.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 1.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 3.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.4 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.7 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 1.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.4 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.8 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 2.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 2.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 3.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 6.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.7 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.9 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 14.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.0 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 10.1 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 1.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.3 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.5 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 1.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 2.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 8.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.4 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 3.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 15.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.8 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 4.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 17.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 3.0 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 2.8 | GO:0070717 | pre-mRNA binding(GO:0036002) poly-purine tract binding(GO:0070717) |
| 0.1 | 4.2 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 4.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.7 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 2.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 2.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 2.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.9 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 4.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.9 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 2.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.5 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 4.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.5 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 3.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 10.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 3.9 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.7 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 5.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0004077 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.0 | 2.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 2.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.5 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 1.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 6.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 20.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 5.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 2.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 4.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 8.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 2.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 5.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 8.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 7.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 2.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 10.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 6.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 7.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 8.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 4.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 4.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 2.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 4.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 4.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 7.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 6.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 4.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 14.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 5.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 25.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 6.0 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.4 | 6.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 13.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 1.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.3 | 4.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.3 | 7.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.3 | 4.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 5.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 3.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 4.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 19.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 5.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 1.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 6.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 8.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 4.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 3.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 7.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 3.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 10.0 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.1 | 2.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 2.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 4.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 7.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 1.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 8.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 5.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 9.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 4.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 4.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 2.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.3 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 2.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 3.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.9 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.9 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |