Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for HSF2
Z-value: 1.02
Transcription factors associated with HSF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF2
|
ENSG00000025156.13 | HSF2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF2 | hg38_v1_chr6_+_122399536_122399568, hg38_v1_chr6_+_122399621_122399692 | 0.07 | 2.8e-01 | Click! |
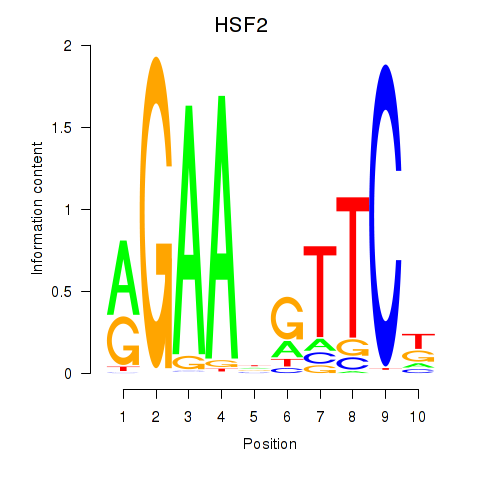
Activity profile of HSF2 motif
Sorted Z-values of HSF2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_76302665 | 20.50 |
ENST00000248553.7
ENST00000674638.1 ENST00000674547.1 ENST00000675226.1 ENST00000675538.1 ENST00000676231.1 ENST00000675134.1 ENST00000675906.1 ENST00000674650.1 |
HSPB1
|
heat shock protein family B (small) member 1 |
| chr18_-_812230 | 11.66 |
ENST00000314574.5
|
YES1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr11_-_111910830 | 9.69 |
ENST00000526167.5
ENST00000651650.1 |
CRYAB
|
crystallin alpha B |
| chr11_-_111910888 | 9.67 |
ENST00000525823.1
ENST00000528961.6 |
CRYAB
|
crystallin alpha B |
| chr6_+_151325665 | 9.51 |
ENST00000354675.10
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr11_-_111910790 | 9.50 |
ENST00000533280.6
|
CRYAB
|
crystallin alpha B |
| chr11_+_75562056 | 7.72 |
ENST00000533603.5
|
SERPINH1
|
serpin family H member 1 |
| chr12_-_52618559 | 7.69 |
ENST00000305748.7
|
KRT73
|
keratin 73 |
| chr11_+_75562274 | 7.67 |
ENST00000532356.5
ENST00000524558.5 |
SERPINH1
|
serpin family H member 1 |
| chr7_+_130486171 | 7.52 |
ENST00000341441.9
ENST00000416162.7 |
MEST
|
mesoderm specific transcript |
| chr7_+_130486324 | 7.50 |
ENST00000427521.6
ENST00000378576.9 |
MEST
|
mesoderm specific transcript |
| chrX_-_103686687 | 7.17 |
ENST00000441076.7
ENST00000422355.5 ENST00000442614.5 ENST00000451301.5 |
MORF4L2
|
mortality factor 4 like 2 |
| chrX_+_136197039 | 6.61 |
ENST00000370683.6
|
FHL1
|
four and a half LIM domains 1 |
| chrX_+_136197020 | 6.34 |
ENST00000370676.7
|
FHL1
|
four and a half LIM domains 1 |
| chrX_+_136196750 | 6.29 |
ENST00000539015.5
|
FHL1
|
four and a half LIM domains 1 |
| chr7_+_128830399 | 6.27 |
ENST00000325888.13
ENST00000346177.6 |
FLNC
|
filamin C |
| chrX_+_136148440 | 6.01 |
ENST00000627383.2
ENST00000630084.2 |
FHL1
|
four and a half LIM domains 1 |
| chr5_-_147453888 | 5.44 |
ENST00000398514.7
|
DPYSL3
|
dihydropyrimidinase like 3 |
| chr18_+_3451647 | 5.30 |
ENST00000345133.9
ENST00000330513.10 ENST00000549546.5 |
TGIF1
|
TGFB induced factor homeobox 1 |
| chrX_-_103688033 | 5.29 |
ENST00000434230.5
ENST00000418819.5 ENST00000360458.5 |
MORF4L2
|
mortality factor 4 like 2 |
| chr6_+_31815532 | 5.15 |
ENST00000375651.7
ENST00000608703.1 |
HSPA1A
|
heat shock protein family A (Hsp70) member 1A |
| chr10_+_91162958 | 5.12 |
ENST00000614189.4
|
PCGF5
|
polycomb group ring finger 5 |
| chr18_+_3451585 | 5.09 |
ENST00000551541.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chrX_-_103688090 | 5.08 |
ENST00000433176.6
|
MORF4L2
|
mortality factor 4 like 2 |
| chr2_+_197500371 | 5.07 |
ENST00000409468.1
ENST00000233893.10 |
HSPE1
|
heat shock protein family E (Hsp10) member 1 |
| chr14_-_52069228 | 5.00 |
ENST00000617139.4
|
NID2
|
nidogen 2 |
| chr8_-_140921188 | 4.95 |
ENST00000517887.5
|
PTK2
|
protein tyrosine kinase 2 |
| chr3_+_159069252 | 4.77 |
ENST00000640015.1
ENST00000476809.7 ENST00000485419.7 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr15_+_96325935 | 4.59 |
ENST00000421109.6
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr9_+_33025265 | 4.58 |
ENST00000330899.5
|
DNAJA1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr12_+_69585434 | 4.40 |
ENST00000299300.11
ENST00000544368.6 |
CCT2
|
chaperonin containing TCP1 subunit 2 |
| chr14_-_52069039 | 4.33 |
ENST00000216286.10
|
NID2
|
nidogen 2 |
| chr2_+_197500398 | 4.29 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr11_-_83285965 | 4.22 |
ENST00000529073.5
ENST00000529611.5 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr2_-_197499857 | 4.13 |
ENST00000428204.6
ENST00000678170.1 ENST00000676933.1 ENST00000678621.1 |
HSPD1
|
heat shock protein family D (Hsp60) member 1 |
| chr7_-_80919017 | 4.01 |
ENST00000265361.8
|
SEMA3C
|
semaphorin 3C |
| chr2_-_197499826 | 3.86 |
ENST00000439605.2
ENST00000388968.8 ENST00000418022.2 |
HSPD1
|
heat shock protein family D (Hsp60) member 1 |
| chr16_+_715092 | 3.86 |
ENST00000568223.7
|
METRN
|
meteorin, glial cell differentiation regulator |
| chr12_+_119178953 | 3.82 |
ENST00000674542.1
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr2_-_187554351 | 3.78 |
ENST00000437725.5
ENST00000409676.5 ENST00000233156.9 ENST00000339091.8 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor |
| chr2_-_177263522 | 3.75 |
ENST00000448782.5
ENST00000446151.6 |
NFE2L2
|
nuclear factor, erythroid 2 like 2 |
| chr12_+_69585666 | 3.67 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1 subunit 2 |
| chr12_+_119178920 | 3.65 |
ENST00000281938.7
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr19_-_14518383 | 3.64 |
ENST00000254322.3
ENST00000595139.2 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr8_+_133113483 | 3.62 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr9_+_89311187 | 3.58 |
ENST00000314355.7
|
CKS2
|
CDC28 protein kinase regulatory subunit 2 |
| chr5_-_73448769 | 3.58 |
ENST00000615637.3
|
FOXD1
|
forkhead box D1 |
| chr9_-_33025052 | 3.56 |
ENST00000673248.1
|
APTX
|
aprataxin |
| chr14_-_102087034 | 3.53 |
ENST00000216281.13
ENST00000553585.5 |
HSP90AA1
|
heat shock protein 90 alpha family class A member 1 |
| chr11_-_27722021 | 3.46 |
ENST00000314915.6
|
BDNF
|
brain derived neurotrophic factor |
| chr6_+_31827730 | 3.31 |
ENST00000375650.5
|
HSPA1B
|
heat shock protein family A (Hsp70) member 1B |
| chr15_+_42359577 | 3.18 |
ENST00000357568.8
|
CAPN3
|
calpain 3 |
| chr11_-_83286328 | 3.10 |
ENST00000525503.5
|
CCDC90B
|
coiled-coil domain containing 90B |
| chr11_-_83286377 | 3.09 |
ENST00000455220.6
ENST00000529689.5 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr9_-_86100123 | 3.02 |
ENST00000388711.7
ENST00000466178.1 |
GOLM1
|
golgi membrane protein 1 |
| chr7_+_157336961 | 2.92 |
ENST00000429029.6
|
DNAJB6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr2_-_10837977 | 2.83 |
ENST00000404824.2
|
PDIA6
|
protein disulfide isomerase family A member 6 |
| chr10_-_99913971 | 2.67 |
ENST00000543621.6
|
DNMBP
|
dynamin binding protein |
| chr3_+_172040554 | 2.63 |
ENST00000336824.8
ENST00000423424.5 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr7_+_157336988 | 2.56 |
ENST00000262177.9
ENST00000417758.5 ENST00000443280.5 |
DNAJB6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chrX_-_153724343 | 2.56 |
ENST00000442093.5
ENST00000345046.12 ENST00000645377.1 ENST00000672675.1 ENST00000647529.1 ENST00000429550.5 |
BCAP31
|
B cell receptor associated protein 31 |
| chr11_-_123062335 | 2.48 |
ENST00000453788.6
ENST00000527387.5 |
HSPA8
|
heat shock protein family A (Hsp70) member 8 |
| chr14_+_88594406 | 2.48 |
ENST00000555900.5
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chrX_-_153724044 | 2.47 |
ENST00000423827.5
ENST00000458587.8 |
BCAP31
|
B cell receptor associated protein 31 |
| chr11_+_66002754 | 2.47 |
ENST00000527348.1
|
BANF1
|
BAF nuclear assembly factor 1 |
| chr14_+_88594430 | 2.38 |
ENST00000406216.7
ENST00000557737.1 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr11_-_65557740 | 2.27 |
ENST00000526927.5
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr11_-_90223036 | 2.25 |
ENST00000320585.11
|
CHORDC1
|
cysteine and histidine rich domain containing 1 |
| chr2_-_216695540 | 2.24 |
ENST00000233813.5
|
IGFBP5
|
insulin like growth factor binding protein 5 |
| chr20_+_4686320 | 2.11 |
ENST00000430350.2
|
PRNP
|
prion protein |
| chr14_+_88594395 | 2.10 |
ENST00000318308.10
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr9_-_33025088 | 2.07 |
ENST00000436040.7
|
APTX
|
aprataxin |
| chr11_-_90223059 | 2.07 |
ENST00000457199.6
ENST00000530765.5 |
CHORDC1
|
cysteine and histidine rich domain containing 1 |
| chr13_-_31161927 | 2.05 |
ENST00000380405.7
|
HSPH1
|
heat shock protein family H (Hsp110) member 1 |
| chr21_+_34073569 | 1.99 |
ENST00000399312.3
ENST00000381151.5 ENST00000362077.4 |
MRPS6
SLC5A3
ENSG00000272657.1
|
mitochondrial ribosomal protein S6 solute carrier family 5 member 3 novel transcript |
| chr20_+_31547367 | 1.93 |
ENST00000394552.3
|
MCTS2P
|
MCTS family member 2, pseudogene |
| chr4_-_158723355 | 1.80 |
ENST00000307720.4
|
PPID
|
peptidylprolyl isomerase D |
| chr11_+_27040725 | 1.79 |
ENST00000529202.5
ENST00000263182.8 |
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr6_-_22297028 | 1.73 |
ENST00000306482.2
|
PRL
|
prolactin |
| chr7_-_26200734 | 1.69 |
ENST00000354667.8
ENST00000618183.5 |
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr11_+_66002225 | 1.67 |
ENST00000445560.6
ENST00000530204.1 |
BANF1
|
BAF nuclear assembly factor 1 |
| chr3_-_48595267 | 1.63 |
ENST00000328333.12
ENST00000681320.1 |
COL7A1
|
collagen type VII alpha 1 chain |
| chr3_-_15797930 | 1.59 |
ENST00000683139.1
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr2_-_187554473 | 1.57 |
ENST00000453013.5
ENST00000417013.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr12_+_2794961 | 1.56 |
ENST00000001008.6
|
FKBP4
|
FKBP prolyl isomerase 4 |
| chr13_-_31161890 | 1.54 |
ENST00000320027.10
|
HSPH1
|
heat shock protein family H (Hsp110) member 1 |
| chr18_+_24139013 | 1.52 |
ENST00000399481.6
ENST00000327201.10 |
CABYR
|
calcium binding tyrosine phosphorylation regulated |
| chr11_-_123062022 | 1.52 |
ENST00000532182.5
ENST00000524590.5 ENST00000528292.5 ENST00000533540.5 ENST00000534624.6 ENST00000525463.5 |
HSPA8
|
heat shock protein family A (Hsp70) member 8 |
| chr11_+_86302211 | 1.49 |
ENST00000533986.5
ENST00000278483.8 |
HIKESHI
|
heat shock protein nuclear import factor hikeshi |
| chr3_+_45026296 | 1.39 |
ENST00000296130.5
|
CLEC3B
|
C-type lectin domain family 3 member B |
| chr11_+_66002475 | 1.37 |
ENST00000312175.7
ENST00000533166.5 |
BANF1
|
BAF nuclear assembly factor 1 |
| chr4_-_152679984 | 1.37 |
ENST00000304385.8
ENST00000504064.1 |
TMEM154
|
transmembrane protein 154 |
| chr6_+_44247866 | 1.36 |
ENST00000371554.2
|
HSP90AB1
|
heat shock protein 90 alpha family class B member 1 |
| chr3_-_15798184 | 1.34 |
ENST00000624145.3
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr8_-_80171106 | 1.25 |
ENST00000519303.6
|
TPD52
|
tumor protein D52 |
| chr10_-_103153609 | 1.24 |
ENST00000675985.1
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr15_-_58065703 | 1.24 |
ENST00000249750.9
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr13_-_31162341 | 1.23 |
ENST00000445273.6
ENST00000630972.2 |
HSPH1
|
heat shock protein family H (Hsp110) member 1 |
| chr1_+_151198536 | 1.21 |
ENST00000349792.9
ENST00000409426.5 ENST00000368888.9 ENST00000441902.6 ENST00000368890.8 ENST00000424999.1 |
PIP5K1A
|
phosphatidylinositol-4-phosphate 5-kinase type 1 alpha |
| chr16_+_71358713 | 1.20 |
ENST00000349553.9
ENST00000302628.9 ENST00000562305.5 |
CALB2
|
calbindin 2 |
| chr6_+_32154131 | 1.19 |
ENST00000375143.6
ENST00000324816.11 ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr11_+_59511368 | 1.17 |
ENST00000641278.1
|
OR4D9
|
olfactory receptor family 4 subfamily D member 9 |
| chrX_-_119606412 | 1.14 |
ENST00000304449.8
|
NKRF
|
NFKB repressing factor |
| chr11_+_66509079 | 1.14 |
ENST00000419755.3
|
ENSG00000256349.1
|
novel protein |
| chr4_-_73223082 | 1.11 |
ENST00000509867.6
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr6_+_32154010 | 1.09 |
ENST00000375137.6
|
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr9_+_841691 | 1.06 |
ENST00000382276.8
|
DMRT1
|
doublesex and mab-3 related transcription factor 1 |
| chr3_-_15440560 | 1.06 |
ENST00000595627.5
ENST00000597949.1 ENST00000494875.3 ENST00000595975.1 ENST00000598878.1 |
EAF1-AS1
METTL6
|
EAF1 antisense RNA 1 methyltransferase like 6 |
| chr1_-_6393750 | 1.04 |
ENST00000545482.5
ENST00000361521.9 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr11_+_12674397 | 1.02 |
ENST00000527636.7
|
TEAD1
|
TEA domain transcription factor 1 |
| chr4_+_76011171 | 0.97 |
ENST00000513353.5
ENST00000341029.9 |
ART3
|
ADP-ribosyltransferase 3 (inactive) |
| chr9_-_13279407 | 0.96 |
ENST00000546205.5
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr2_-_85354500 | 0.87 |
ENST00000449375.1
ENST00000409984.2 ENST00000295802.9 |
RETSAT
|
retinol saturase |
| chr2_-_96740034 | 0.82 |
ENST00000264963.9
ENST00000377079.8 |
LMAN2L
|
lectin, mannose binding 2 like |
| chr1_-_120176450 | 0.81 |
ENST00000578049.4
|
SEC22B
|
SEC22 homolog B, vesicle trafficking protein |
| chr15_-_43493105 | 0.79 |
ENST00000382039.7
ENST00000450115.6 ENST00000382044.9 |
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr6_+_146543824 | 0.79 |
ENST00000367495.4
|
RAB32
|
RAB32, member RAS oncogene family |
| chr6_+_32153441 | 0.77 |
ENST00000414204.5
ENST00000361568.6 ENST00000395523.5 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr11_+_59511539 | 0.76 |
ENST00000641962.1
|
OR4D9
|
olfactory receptor family 4 subfamily D member 9 |
| chr21_-_29073565 | 0.75 |
ENST00000431234.1
ENST00000286788.9 ENST00000540844.5 |
CCT8
|
chaperonin containing TCP1 subunit 8 |
| chr5_+_80035341 | 0.72 |
ENST00000350881.6
|
THBS4
|
thrombospondin 4 |
| chr17_-_58417521 | 0.71 |
ENST00000584437.5
ENST00000407977.7 |
RNF43
|
ring finger protein 43 |
| chr11_+_72080803 | 0.67 |
ENST00000423494.6
ENST00000539587.6 ENST00000536917.2 ENST00000538478.5 ENST00000324866.11 ENST00000643715.1 ENST00000439209.5 |
LRTOMT
ENSG00000284922.2
|
leucine rich transmembrane and O-methyltransferase domain containing leucine rich transmembrane and O-methyltransferase domain containing |
| chr11_+_72080595 | 0.66 |
ENST00000647530.1
ENST00000539271.6 ENST00000642510.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr9_-_34710069 | 0.66 |
ENST00000378792.1
ENST00000259607.7 |
CCL21
|
C-C motif chemokine ligand 21 |
| chr1_-_168729187 | 0.66 |
ENST00000367817.4
|
DPT
|
dermatopontin |
| chr1_-_6393339 | 0.65 |
ENST00000608083.5
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr6_-_53148822 | 0.65 |
ENST00000259803.8
|
GCM1
|
glial cells missing transcription factor 1 |
| chr20_+_63235899 | 0.60 |
ENST00000217169.8
|
BIRC7
|
baculoviral IAP repeat containing 7 |
| chr8_+_109540075 | 0.59 |
ENST00000614147.1
ENST00000337573.10 |
EBAG9
|
estrogen receptor binding site associated antigen 9 |
| chr4_-_107036302 | 0.58 |
ENST00000285311.8
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr6_-_155314444 | 0.51 |
ENST00000367166.5
|
TFB1M
|
transcription factor B1, mitochondrial |
| chr17_-_58417547 | 0.50 |
ENST00000577716.5
|
RNF43
|
ring finger protein 43 |
| chr6_-_32371872 | 0.47 |
ENST00000527965.5
ENST00000532023.5 ENST00000447241.6 ENST00000534588.1 |
TSBP1
|
testis expressed basic protein 1 |
| chr13_+_49110309 | 0.44 |
ENST00000398316.7
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr8_-_119592954 | 0.43 |
ENST00000522167.5
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr20_+_46118300 | 0.43 |
ENST00000372285.8
ENST00000372276.7 |
CD40
|
CD40 molecule |
| chr1_-_197775435 | 0.38 |
ENST00000620048.6
|
DENND1B
|
DENN domain containing 1B |
| chrX_-_19965142 | 0.38 |
ENST00000340625.3
|
BCLAF3
|
BCLAF1 and THRAP3 family member 3 |
| chr7_-_42152396 | 0.35 |
ENST00000642432.1
ENST00000647255.1 ENST00000677288.1 |
GLI3
|
GLI family zinc finger 3 |
| chr2_-_218286763 | 0.34 |
ENST00000444881.5
|
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr3_+_14947568 | 0.31 |
ENST00000413118.5
ENST00000425241.5 |
NR2C2
|
nuclear receptor subfamily 2 group C member 2 |
| chr6_-_142946312 | 0.28 |
ENST00000367604.6
|
HIVEP2
|
HIVEP zinc finger 2 |
| chr18_-_46917432 | 0.25 |
ENST00000324794.11
ENST00000545673.5 ENST00000585916.6 |
PIAS2
|
protein inhibitor of activated STAT 2 |
| chr7_-_42152444 | 0.23 |
ENST00000479210.1
|
GLI3
|
GLI family zinc finger 3 |
| chr6_-_32371912 | 0.20 |
ENST00000612031.4
|
TSBP1
|
testis expressed basic protein 1 |
| chr5_-_149063021 | 0.18 |
ENST00000515425.6
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr6_-_32371897 | 0.17 |
ENST00000442822.6
ENST00000375015.8 ENST00000533191.5 |
TSBP1
|
testis expressed basic protein 1 |
| chr6_+_29396555 | 0.16 |
ENST00000623183.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2 |
| chr8_-_80171496 | 0.16 |
ENST00000379096.9
ENST00000518937.6 |
TPD52
|
tumor protein D52 |
| chr6_-_135054785 | 0.15 |
ENST00000367820.6
ENST00000314674.7 ENST00000524715.5 ENST00000415177.6 ENST00000367826.6 ENST00000367837.10 |
HBS1L
|
HBS1 like translational GTPase |
| chr17_-_41505597 | 0.14 |
ENST00000336861.7
ENST00000246635.8 ENST00000587544.5 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr3_-_129688691 | 0.13 |
ENST00000432054.6
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr2_+_11542662 | 0.11 |
ENST00000389825.7
ENST00000381483.6 |
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr1_+_26921715 | 0.09 |
ENST00000321265.10
|
NUDC
|
nuclear distribution C, dynein complex regulator |
| chr20_+_63235883 | 0.07 |
ENST00000342412.10
|
BIRC7
|
baculoviral IAP repeat containing 7 |
| chr1_-_184037695 | 0.05 |
ENST00000361927.9
ENST00000649786.1 |
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr6_-_109094819 | 0.03 |
ENST00000436639.6
|
SESN1
|
sestrin 1 |
| chr5_-_22853320 | 0.00 |
ENST00000504376.6
ENST00000382254.6 |
CDH12
|
cadherin 12 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 20.5 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 2.7 | 8.0 | GO:0002368 | B cell cytokine production(GO:0002368) |
| 2.1 | 8.5 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 1.9 | 15.4 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 1.7 | 28.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.6 | 8.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 1.4 | 4.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 1.3 | 5.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.2 | 3.5 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 1.1 | 4.6 | GO:0009956 | radial pattern formation(GO:0009956) |
| 1.0 | 9.4 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.9 | 3.8 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.9 | 3.5 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.8 | 4.0 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.8 | 9.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.8 | 2.3 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.7 | 2.2 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.7 | 3.6 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.7 | 5.5 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.7 | 4.0 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.7 | 2.0 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.6 | 17.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.6 | 5.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.6 | 5.6 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.5 | 8.7 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.5 | 2.1 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 0.5 | 3.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.5 | 1.4 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.4 | 7.0 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.4 | 1.7 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.4 | 11.7 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.4 | 1.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.4 | 5.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.4 | 1.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.4 | 3.2 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.4 | 4.9 | GO:0051964 | netrin-activated signaling pathway(GO:0038007) negative regulation of synapse assembly(GO:0051964) |
| 0.3 | 24.3 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.3 | 3.0 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.3 | 9.3 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.2 | 1.7 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 0.7 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.2 | 5.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 1.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 0.7 | GO:0097026 | mesangial cell-matrix adhesion(GO:0035759) dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.2 | 0.6 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.2 | 1.8 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 0.6 | GO:0060366 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.2 | 1.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 1.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.8 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.8 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 9.8 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.1 | 6.3 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) negative regulation of autophagosome assembly(GO:1902902) vesicle fusion with endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membrane(GO:1990668) |
| 0.1 | 3.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 15.0 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.1 | 0.7 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) |
| 0.1 | 1.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 1.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.3 | GO:0060117 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) auditory receptor cell development(GO:0060117) |
| 0.1 | 0.3 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 7.8 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 4.6 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.7 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.7 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 1.0 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 3.9 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 2.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.2 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.3 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 1.0 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.4 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.5 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 2.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 28.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.9 | 8.0 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) lipopolysaccharide receptor complex(GO:0046696) |
| 0.8 | 5.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.6 | 8.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.5 | 17.5 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.5 | 1.5 | GO:0097229 | sperm end piece(GO:0097229) |
| 0.5 | 4.0 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.5 | 4.6 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.5 | 1.4 | GO:1990913 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 0.3 | 1.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.3 | 20.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.3 | 7.7 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.3 | 2.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 1.4 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 2.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 7.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 8.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 2.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 0.8 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 17.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 6.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 5.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 5.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 1.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 9.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 2.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 3.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 46.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 8.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.0 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 1.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 5.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 6.3 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 3.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 11.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 3.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 3.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.5 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 1.9 | 5.6 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 1.5 | 16.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 1.3 | 8.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 1.0 | 28.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.9 | 3.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.6 | 3.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.6 | 1.8 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.5 | 5.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.5 | 9.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.5 | 1.4 | GO:0002135 | CTP binding(GO:0002135) |
| 0.4 | 1.7 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.4 | 2.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.4 | 1.2 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.4 | 1.2 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.4 | 3.5 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.4 | 1.6 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.4 | 4.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 10.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.3 | 2.0 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.3 | 36.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.3 | 3.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.3 | 0.8 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.3 | 4.9 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 4.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 6.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 1.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 11.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 1.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 3.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 2.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 7.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 5.0 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 0.8 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 1.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 2.8 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 1.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 4.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 1.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.4 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 23.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 3.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 4.8 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 4.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 8.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 7.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 1.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 5.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 3.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 20.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 11.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 8.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 8.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 10.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 3.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 3.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 6.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 11.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 4.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 15.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 3.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 5.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.8 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 4.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.7 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.5 | 5.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 27.8 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.3 | 8.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.3 | 4.9 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.3 | 17.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 4.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 5.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 10.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 4.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 8.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 5.3 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 2.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 5.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 4.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |