Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for HSF4
Z-value: 10.60
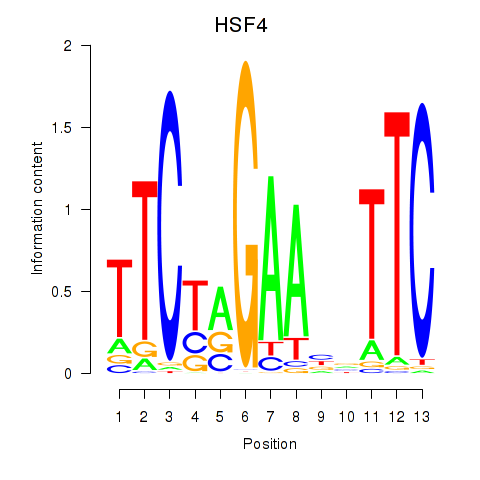
Transcription factors associated with HSF4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF4
|
ENSG00000102878.18 | HSF4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF4 | hg38_v1_chr16_+_67164730_67164768 | -0.68 | 5.3e-31 | Click! |
Activity profile of HSF4 motif
Sorted Z-values of HSF4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_56051685 | 94.07 |
ENST00000335503.3
|
CCT6A
|
chaperonin containing TCP1 subunit 6A |
| chr7_+_56051756 | 82.29 |
ENST00000275603.9
|
CCT6A
|
chaperonin containing TCP1 subunit 6A |
| chr17_-_42016680 | 78.89 |
ENST00000674497.1
|
DNAJC7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr11_+_64186219 | 69.10 |
ENST00000543847.1
|
STIP1
|
stress induced phosphoprotein 1 |
| chr9_+_33025265 | 60.79 |
ENST00000330899.5
|
DNAJA1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr13_-_31161927 | 58.69 |
ENST00000380405.7
|
HSPH1
|
heat shock protein family H (Hsp110) member 1 |
| chr14_-_102087034 | 58.03 |
ENST00000216281.13
ENST00000553585.5 |
HSP90AA1
|
heat shock protein 90 alpha family class A member 1 |
| chr11_-_123062335 | 55.40 |
ENST00000453788.6
ENST00000527387.5 |
HSPA8
|
heat shock protein family A (Hsp70) member 8 |
| chr11_-_123062022 | 54.11 |
ENST00000532182.5
ENST00000524590.5 ENST00000528292.5 ENST00000533540.5 ENST00000534624.6 ENST00000525463.5 |
HSPA8
|
heat shock protein family A (Hsp70) member 8 |
| chr12_+_2794961 | 52.75 |
ENST00000001008.6
|
FKBP4
|
FKBP prolyl isomerase 4 |
| chr9_-_86100123 | 52.24 |
ENST00000388711.7
ENST00000466178.1 |
GOLM1
|
golgi membrane protein 1 |
| chr2_+_200306048 | 51.23 |
ENST00000409988.7
ENST00000409385.5 |
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr11_+_64186163 | 50.88 |
ENST00000305218.9
ENST00000538945.5 |
STIP1
|
stress induced phosphoprotein 1 |
| chr2_+_200306519 | 48.87 |
ENST00000360760.9
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chrX_-_103688033 | 48.78 |
ENST00000434230.5
ENST00000418819.5 ENST00000360458.5 |
MORF4L2
|
mortality factor 4 like 2 |
| chrX_-_103688090 | 47.20 |
ENST00000433176.6
|
MORF4L2
|
mortality factor 4 like 2 |
| chr13_-_31161890 | 46.61 |
ENST00000320027.10
|
HSPH1
|
heat shock protein family H (Hsp110) member 1 |
| chr21_-_29073565 | 46.59 |
ENST00000431234.1
ENST00000286788.9 ENST00000540844.5 |
CCT8
|
chaperonin containing TCP1 subunit 8 |
| chr1_-_45521854 | 45.11 |
ENST00000372079.1
ENST00000319248.13 |
PRDX1
|
peroxiredoxin 1 |
| chr2_-_61888570 | 43.34 |
ENST00000394440.8
ENST00000544079.2 |
CCT4
|
chaperonin containing TCP1 subunit 4 |
| chr1_+_154974672 | 41.83 |
ENST00000308987.6
ENST00000368436.1 |
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr1_-_45521931 | 41.72 |
ENST00000447184.6
ENST00000262746.5 |
PRDX1
|
peroxiredoxin 1 |
| chr13_-_31162341 | 41.49 |
ENST00000445273.6
ENST00000630972.2 |
HSPH1
|
heat shock protein family H (Hsp110) member 1 |
| chr6_+_44246166 | 39.02 |
ENST00000620073.4
|
HSP90AB1
|
heat shock protein 90 alpha family class B member 1 |
| chr9_-_83978429 | 38.94 |
ENST00000351839.7
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr9_-_86099506 | 37.40 |
ENST00000388712.7
|
GOLM1
|
golgi membrane protein 1 |
| chr11_+_75562056 | 37.39 |
ENST00000533603.5
|
SERPINH1
|
serpin family H member 1 |
| chr6_+_44247866 | 37.07 |
ENST00000371554.2
|
HSP90AB1
|
heat shock protein 90 alpha family class B member 1 |
| chr11_+_75562242 | 37.07 |
ENST00000526397.5
ENST00000529643.1 ENST00000525492.5 ENST00000530284.5 ENST00000358171.8 |
SERPINH1
|
serpin family H member 1 |
| chr1_+_154974653 | 36.23 |
ENST00000368439.5
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr2_-_69437588 | 36.04 |
ENST00000394305.5
|
NFU1
|
NFU1 iron-sulfur cluster scaffold |
| chr12_+_103930600 | 35.89 |
ENST00000680316.1
ENST00000679861.1 |
HSP90B1
|
heat shock protein 90 beta family member 1 |
| chr2_+_197500398 | 35.28 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr1_-_244863085 | 35.22 |
ENST00000440865.2
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U |
| chr11_-_83285965 | 35.14 |
ENST00000529073.5
ENST00000529611.5 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr7_+_76303547 | 34.87 |
ENST00000429938.1
|
HSPB1
|
heat shock protein family B (small) member 1 |
| chr12_-_110445540 | 34.02 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex subunit 3 |
| chr4_-_17512079 | 33.96 |
ENST00000428702.6
ENST00000508623.5 ENST00000513615.5 ENST00000281243.10 |
QDPR
|
quinoid dihydropteridine reductase |
| chr11_+_66002754 | 33.92 |
ENST00000527348.1
|
BANF1
|
BAF nuclear assembly factor 1 |
| chr11_-_83286328 | 33.10 |
ENST00000525503.5
|
CCDC90B
|
coiled-coil domain containing 90B |
| chr5_+_10250216 | 32.98 |
ENST00000515390.5
|
CCT5
|
chaperonin containing TCP1 subunit 5 |
| chr11_+_66002225 | 32.97 |
ENST00000445560.6
ENST00000530204.1 |
BANF1
|
BAF nuclear assembly factor 1 |
| chr12_+_103930332 | 32.95 |
ENST00000681861.1
ENST00000550595.2 ENST00000680762.1 ENST00000614327.2 ENST00000681949.1 ENST00000299767.10 |
HSP90B1
|
heat shock protein 90 beta family member 1 |
| chr12_+_69585434 | 32.94 |
ENST00000299300.11
ENST00000544368.6 |
CCT2
|
chaperonin containing TCP1 subunit 2 |
| chr9_-_33025052 | 32.90 |
ENST00000673248.1
|
APTX
|
aprataxin |
| chr2_-_197499826 | 32.82 |
ENST00000439605.2
ENST00000388968.8 ENST00000418022.2 |
HSPD1
|
heat shock protein family D (Hsp60) member 1 |
| chr7_-_26200734 | 32.65 |
ENST00000354667.8
ENST00000618183.5 |
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr9_-_33025088 | 32.31 |
ENST00000436040.7
|
APTX
|
aprataxin |
| chr11_+_75562274 | 31.97 |
ENST00000532356.5
ENST00000524558.5 |
SERPINH1
|
serpin family H member 1 |
| chr11_-_83286377 | 31.56 |
ENST00000455220.6
ENST00000529689.5 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr7_+_76302665 | 31.53 |
ENST00000248553.7
ENST00000674638.1 ENST00000674547.1 ENST00000675226.1 ENST00000675538.1 ENST00000676231.1 ENST00000675134.1 ENST00000675906.1 ENST00000674650.1 |
HSPB1
|
heat shock protein family B (small) member 1 |
| chr2_+_197500371 | 30.97 |
ENST00000409468.1
ENST00000233893.10 |
HSPE1
|
heat shock protein family E (Hsp10) member 1 |
| chr2_-_197499857 | 30.21 |
ENST00000428204.6
ENST00000678170.1 ENST00000676933.1 ENST00000678621.1 |
HSPD1
|
heat shock protein family D (Hsp60) member 1 |
| chrX_-_153724044 | 29.97 |
ENST00000423827.5
ENST00000458587.8 |
BCAP31
|
B cell receptor associated protein 31 |
| chrX_-_153724343 | 29.94 |
ENST00000442093.5
ENST00000345046.12 ENST00000645377.1 ENST00000672675.1 ENST00000647529.1 ENST00000429550.5 |
BCAP31
|
B cell receptor associated protein 31 |
| chr1_-_6393339 | 29.48 |
ENST00000608083.5
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr5_+_10250272 | 29.45 |
ENST00000280326.9
ENST00000625723.1 |
CCT5
|
chaperonin containing TCP1 subunit 5 |
| chr21_-_39349048 | 29.25 |
ENST00000380748.5
ENST00000380749.10 |
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr11_+_66002475 | 29.24 |
ENST00000312175.7
ENST00000533166.5 |
BANF1
|
BAF nuclear assembly factor 1 |
| chr1_-_6393750 | 29.21 |
ENST00000545482.5
ENST00000361521.9 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr2_+_200306340 | 26.18 |
ENST00000451764.6
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr22_-_40856565 | 25.43 |
ENST00000620312.4
ENST00000216218.8 |
ST13
|
ST13 Hsp70 interacting protein |
| chr7_-_56051544 | 25.27 |
ENST00000395471.7
|
PSPH
|
phosphoserine phosphatase |
| chr1_-_156338226 | 24.82 |
ENST00000496684.6
ENST00000368259.6 ENST00000472765.6 ENST00000295688.8 ENST00000533194.5 ENST00000478640.6 |
CCT3
|
chaperonin containing TCP1 subunit 3 |
| chr1_+_26921715 | 24.29 |
ENST00000321265.10
|
NUDC
|
nuclear distribution C, dynein complex regulator |
| chr1_+_175000126 | 23.64 |
ENST00000406752.1
ENST00000405362.1 |
CACYBP
|
calcyclin binding protein |
| chr17_-_63773534 | 23.63 |
ENST00000403162.7
ENST00000582252.1 ENST00000225726.10 |
CCDC47
|
coiled-coil domain containing 47 |
| chr4_-_158723355 | 23.61 |
ENST00000307720.4
|
PPID
|
peptidylprolyl isomerase D |
| chr3_+_159069252 | 23.08 |
ENST00000640015.1
ENST00000476809.7 ENST00000485419.7 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr16_-_8861713 | 22.63 |
ENST00000567554.5
|
CARHSP1
|
calcium regulated heat stable protein 1 |
| chrX_+_123961304 | 22.62 |
ENST00000371160.5
ENST00000435103.5 |
STAG2
|
stromal antigen 2 |
| chr1_-_6200831 | 22.55 |
ENST00000497965.5
|
RPL22
|
ribosomal protein L22 |
| chr1_-_53238485 | 22.31 |
ENST00000371466.4
ENST00000371470.8 |
MAGOH
|
mago homolog, exon junction complex subunit |
| chr15_+_43800586 | 21.44 |
ENST00000442995.4
ENST00000458412.2 |
HYPK
|
huntingtin interacting protein K |
| chr11_+_20363685 | 21.44 |
ENST00000530266.5
ENST00000451739.7 ENST00000421577.6 ENST00000443524.6 ENST00000419348.6 |
HTATIP2
|
HIV-1 Tat interactive protein 2 |
| chr13_-_30617500 | 21.01 |
ENST00000405805.5
|
HMGB1
|
high mobility group box 1 |
| chr19_-_14518383 | 20.60 |
ENST00000254322.3
ENST00000595139.2 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr4_+_41256921 | 20.08 |
ENST00000284440.9
ENST00000508768.5 ENST00000512788.1 |
UCHL1
|
ubiquitin C-terminal hydrolase L1 |
| chr15_+_96325935 | 19.41 |
ENST00000421109.6
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr16_-_8861799 | 19.02 |
ENST00000610831.4
ENST00000614449.4 |
CARHSP1
|
calcium regulated heat stable protein 1 |
| chr20_+_19886506 | 18.91 |
ENST00000648440.1
|
RIN2
|
Ras and Rab interactor 2 |
| chr3_-_185938006 | 18.83 |
ENST00000342294.4
ENST00000453386.7 ENST00000382191.4 |
TRA2B
|
transformer 2 beta homolog |
| chr14_-_23071617 | 17.41 |
ENST00000357481.6
|
ACIN1
|
apoptotic chromatin condensation inducer 1 |
| chr10_-_73096850 | 17.03 |
ENST00000307116.6
ENST00000373008.6 ENST00000394890.7 |
P4HA1
|
prolyl 4-hydroxylase subunit alpha 1 |
| chr9_-_6015607 | 16.06 |
ENST00000485372.1
ENST00000259569.6 ENST00000623170.1 |
RANBP6
|
RAN binding protein 6 |
| chr14_-_23071538 | 15.90 |
ENST00000555566.1
ENST00000338631.10 ENST00000557515.5 ENST00000397341.7 |
ACIN1
|
apoptotic chromatin condensation inducer 1 |
| chr11_-_90223059 | 15.59 |
ENST00000457199.6
ENST00000530765.5 |
CHORDC1
|
cysteine and histidine rich domain containing 1 |
| chr11_+_86302211 | 15.57 |
ENST00000533986.5
ENST00000278483.8 |
HIKESHI
|
heat shock protein nuclear import factor hikeshi |
| chr11_-_90223036 | 15.30 |
ENST00000320585.11
|
CHORDC1
|
cysteine and histidine rich domain containing 1 |
| chr16_-_8861744 | 15.21 |
ENST00000569398.5
ENST00000568968.1 |
CARHSP1
|
calcium regulated heat stable protein 1 |
| chr11_+_10751203 | 15.16 |
ENST00000361367.7
|
CTR9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr7_-_56051288 | 15.04 |
ENST00000419984.6
ENST00000413218.5 ENST00000275605.8 ENST00000424596.1 ENST00000421312.5 ENST00000416592.1 |
PSPH
|
phosphoserine phosphatase |
| chr19_-_14517425 | 13.86 |
ENST00000676577.1
ENST00000677204.1 ENST00000598235.2 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr20_+_4686320 | 13.75 |
ENST00000430350.2
|
PRNP
|
prion protein |
| chr4_+_41359599 | 13.25 |
ENST00000513024.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_+_49851205 | 12.80 |
ENST00000601675.5
|
PTOV1
|
PTOV1 extended AT-hook containing adaptor protein |
| chr19_+_49851136 | 12.44 |
ENST00000391842.5
|
PTOV1
|
PTOV1 extended AT-hook containing adaptor protein |
| chr7_+_157336961 | 12.09 |
ENST00000429029.6
|
DNAJB6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr8_+_37736612 | 11.99 |
ENST00000518526.5
ENST00000523887.5 ENST00000648919.1 ENST00000519638.3 |
ERLIN2
|
ER lipid raft associated 2 |
| chr19_+_49851173 | 11.83 |
ENST00000599732.5
|
PTOV1
|
PTOV1 extended AT-hook containing adaptor protein |
| chrX_-_107000185 | 11.62 |
ENST00000355610.9
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr19_+_2269520 | 11.43 |
ENST00000602676.6
ENST00000582888.8 |
OAZ1
|
ornithine decarboxylase antizyme 1 |
| chr12_-_48682158 | 11.40 |
ENST00000553086.5
ENST00000548304.1 ENST00000550347.5 ENST00000420613.7 ENST00000550931.5 ENST00000550870.1 |
KANSL2
|
KAT8 regulatory NSL complex subunit 2 |
| chr1_-_25905989 | 11.27 |
ENST00000399728.5
|
STMN1
|
stathmin 1 |
| chr2_+_200305976 | 10.79 |
ENST00000358677.9
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr1_+_174999925 | 10.64 |
ENST00000367679.7
|
CACYBP
|
calcyclin binding protein |
| chr20_-_44960348 | 10.55 |
ENST00000372813.4
|
TOMM34
|
translocase of outer mitochondrial membrane 34 |
| chr6_-_106325416 | 10.34 |
ENST00000343245.7
|
ATG5
|
autophagy related 5 |
| chr6_+_4889992 | 10.32 |
ENST00000343762.5
|
CDYL
|
chromodomain Y like |
| chrX_+_123960519 | 10.31 |
ENST00000455404.5
ENST00000218089.13 |
STAG2
|
stromal antigen 2 |
| chr3_+_130894050 | 10.17 |
ENST00000510168.6
|
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr12_-_124917340 | 10.11 |
ENST00000542416.1
|
UBC
|
ubiquitin C |
| chr5_-_140673568 | 9.99 |
ENST00000542735.2
|
DND1
|
DND microRNA-mediated repression inhibitor 1 |
| chr14_+_71586261 | 9.63 |
ENST00000358550.6
|
SIPA1L1
|
signal induced proliferation associated 1 like 1 |
| chr2_+_85539158 | 9.59 |
ENST00000306434.8
|
MAT2A
|
methionine adenosyltransferase 2A |
| chr2_+_227813834 | 9.49 |
ENST00000358813.5
ENST00000409189.7 |
CCL20
|
C-C motif chemokine ligand 20 |
| chr12_-_122526929 | 9.48 |
ENST00000331738.12
ENST00000528279.1 ENST00000344591.8 ENST00000526560.6 |
RSRC2
|
arginine and serine rich coiled-coil 2 |
| chr22_+_29268248 | 8.67 |
ENST00000436425.5
ENST00000447973.5 ENST00000397938.7 ENST00000406548.5 ENST00000437155.6 ENST00000415761.5 ENST00000331029.11 |
EWSR1
|
EWS RNA binding protein 1 |
| chr1_-_120176450 | 8.38 |
ENST00000578049.4
|
SEC22B
|
SEC22 homolog B, vesicle trafficking protein |
| chr11_-_27700472 | 8.32 |
ENST00000418212.5
ENST00000533246.5 |
BDNF
|
brain derived neurotrophic factor |
| chr5_+_134371561 | 8.02 |
ENST00000265339.7
ENST00000506787.5 ENST00000507277.1 |
UBE2B
|
ubiquitin conjugating enzyme E2 B |
| chr3_+_130894157 | 7.90 |
ENST00000504948.5
ENST00000513801.5 ENST00000505072.5 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr18_+_59899988 | 7.79 |
ENST00000316660.7
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr3_+_130894382 | 7.53 |
ENST00000509662.5
ENST00000328560.12 ENST00000428331.6 ENST00000359644.7 ENST00000422190.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr7_+_157336988 | 7.46 |
ENST00000262177.9
ENST00000417758.5 ENST00000443280.5 |
DNAJB6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr15_+_78872809 | 7.44 |
ENST00000331268.9
|
MORF4L1
|
mortality factor 4 like 1 |
| chr4_-_25863537 | 7.36 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr15_+_78872881 | 7.24 |
ENST00000559930.5
ENST00000426013.7 |
MORF4L1
|
mortality factor 4 like 1 |
| chr16_-_18375069 | 7.22 |
ENST00000545114.5
|
NPIPA9
|
nuclear pore complex interacting protein family, member A9 |
| chr6_-_41941795 | 7.10 |
ENST00000372991.9
|
CCND3
|
cyclin D3 |
| chr11_+_20363780 | 6.78 |
ENST00000532505.1
|
HTATIP2
|
HIV-1 Tat interactive protein 2 |
| chr19_-_46714269 | 6.60 |
ENST00000600194.5
|
PRKD2
|
protein kinase D2 |
| chr3_+_130893959 | 6.42 |
ENST00000508532.5
ENST00000533801.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr5_+_74766981 | 6.33 |
ENST00000296802.9
|
NSA2
|
NSA2 ribosome biogenesis factor |
| chr6_-_41941507 | 6.32 |
ENST00000372987.8
|
CCND3
|
cyclin D3 |
| chr5_+_141223332 | 6.31 |
ENST00000239449.7
ENST00000624896.1 ENST00000624396.1 |
PCDHB14
ENSG00000279983.1
|
protocadherin beta 14 novel protein |
| chr10_-_20897288 | 6.12 |
ENST00000377122.9
|
NEBL
|
nebulette |
| chr5_-_178187364 | 6.01 |
ENST00000463439.3
|
GMCL2
|
germ cell-less 2, spermatogenesis associated |
| chr6_-_41941728 | 6.00 |
ENST00000414200.6
|
CCND3
|
cyclin D3 |
| chr21_+_33403466 | 5.76 |
ENST00000405436.5
|
IFNGR2
|
interferon gamma receptor 2 |
| chr2_+_200306648 | 5.40 |
ENST00000409140.8
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr5_+_446139 | 5.34 |
ENST00000315013.9
|
EXOC3
|
exocyst complex component 3 |
| chr18_-_21704763 | 5.24 |
ENST00000580981.5
ENST00000289119.7 |
ABHD3
|
abhydrolase domain containing 3, phospholipase |
| chr17_-_64230727 | 5.19 |
ENST00000583097.5
ENST00000615733.4 |
TEX2
|
testis expressed 2 |
| chr2_-_47906437 | 4.85 |
ENST00000403359.8
|
FBXO11
|
F-box protein 11 |
| chr2_-_237590660 | 4.02 |
ENST00000409576.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr12_+_65169546 | 3.67 |
ENST00000308330.3
|
LEMD3
|
LEM domain containing 3 |
| chr11_-_47848467 | 3.57 |
ENST00000378460.6
|
NUP160
|
nucleoporin 160 |
| chr18_+_11751467 | 2.63 |
ENST00000535121.5
|
GNAL
|
G protein subunit alpha L |
| chr3_+_186783567 | 2.47 |
ENST00000323963.10
ENST00000440191.6 |
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr19_-_46471484 | 2.33 |
ENST00000313683.15
ENST00000602246.1 |
PNMA8A
|
PNMA family member 8A |
| chr6_+_31715339 | 2.21 |
ENST00000375824.1
ENST00000375825.7 |
LY6G6D
|
lymphocyte antigen 6 family member G6D |
| chr12_+_69738853 | 1.92 |
ENST00000247833.12
ENST00000378815.10 ENST00000483530.6 |
RAB3IP
|
RAB3A interacting protein |
| chr22_+_32475257 | 1.85 |
ENST00000397426.5
|
FBXO7
|
F-box protein 7 |
| chr3_-_195876635 | 1.77 |
ENST00000672669.1
ENST00000672886.1 ENST00000672098.1 ENST00000671767.1 ENST00000672548.1 |
TNK2
|
tyrosine kinase non receptor 2 |
| chr7_+_130380339 | 1.64 |
ENST00000481342.5
ENST00000604896.5 ENST00000011292.8 |
CPA1
|
carboxypeptidase A1 |
| chr7_+_130381092 | 1.41 |
ENST00000484324.1
|
CPA1
|
carboxypeptidase A1 |
| chr17_+_42017020 | 1.39 |
ENST00000307641.9
|
NKIRAS2
|
NFKB inhibitor interacting Ras like 2 |
| chr11_+_20364119 | 1.18 |
ENST00000532081.1
ENST00000531058.1 |
HTATIP2
|
HIV-1 Tat interactive protein 2 |
| chr17_+_34319427 | 0.51 |
ENST00000394620.2
|
CCL8
|
C-C motif chemokine ligand 8 |
| chr18_+_11751494 | 0.44 |
ENST00000269162.9
|
GNAL
|
G protein subunit alpha L |
| chr12_-_57111338 | 0.31 |
ENST00000538913.6
ENST00000537215.6 ENST00000300134.8 ENST00000454075.7 ENST00000640254.2 ENST00000553275.1 ENST00000553533.2 |
STAT6
|
signal transducer and activator of transcription 6 |
| chr19_-_18538371 | 0.20 |
ENST00000596015.1
|
FKBP8
|
FKBP prolyl isomerase 8 |
| chr12_-_56636318 | 0.09 |
ENST00000549506.5
ENST00000379441.7 ENST00000551812.5 |
BAZ2A
|
bromodomain adjacent to zinc finger domain 2A |
| chr1_-_24143112 | 0.08 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 27.6 | 386.5 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 25.4 | 76.1 | GO:1901388 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 23.1 | 207.6 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 21.9 | 109.5 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 21.0 | 63.0 | GO:0002368 | B cell cytokine production(GO:0002368) |
| 16.6 | 66.4 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 15.0 | 59.9 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 14.7 | 58.7 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 14.5 | 58.0 | GO:0043335 | protein unfolding(GO:0043335) |
| 13.3 | 106.4 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 10.3 | 30.9 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 9.7 | 38.9 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 8.2 | 138.8 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 6.9 | 96.1 | GO:0015074 | DNA integration(GO:0015074) |
| 6.5 | 65.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 5.3 | 32.0 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 5.3 | 21.0 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 5.1 | 15.2 | GO:2001168 | histone H3-K79 methylation(GO:0034729) regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 4.9 | 19.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 4.7 | 32.7 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 4.7 | 32.6 | GO:0010044 | response to aluminum ion(GO:0010044) |
| 4.3 | 68.8 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 4.1 | 110.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 4.0 | 20.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 3.7 | 33.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 3.4 | 10.3 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 3.4 | 13.7 | GO:1990535 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 3.2 | 9.5 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 3.1 | 86.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 3.1 | 40.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 3.1 | 31.0 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 2.8 | 64.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 2.8 | 8.3 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 2.7 | 32.9 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 2.7 | 8.0 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of meiosis I(GO:0060903) |
| 2.6 | 23.6 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 2.5 | 35.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 2.4 | 19.6 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 2.4 | 9.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 2.4 | 23.6 | GO:0006983 | ER overload response(GO:0006983) |
| 2.3 | 11.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 2.2 | 97.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 2.1 | 36.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 2.0 | 12.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 1.9 | 17.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 1.4 | 18.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 1.3 | 4.0 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 1.1 | 16.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.0 | 8.4 | GO:1990668 | vesicle fusion with endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membrane(GO:1990668) |
| 0.9 | 7.8 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.8 | 29.3 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.7 | 34.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.7 | 3.7 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.7 | 6.6 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.6 | 11.4 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.5 | 5.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.5 | 10.0 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.5 | 10.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.3 | 4.9 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.3 | 22.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.3 | 34.3 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.3 | 6.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.3 | 58.5 | GO:0006997 | nucleus organization(GO:0006997) |
| 0.3 | 56.9 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.2 | 6.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.2 | 20.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 24.3 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.2 | 6.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 5.8 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 9.6 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 15.6 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 1.8 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.1 | 3.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 2.2 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.3 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 2.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 10.3 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.1 | 10.0 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.1 | 13.3 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 13.8 | GO:0051170 | nuclear import(GO:0051170) |
| 0.0 | 1.8 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 25.4 | 76.1 | GO:1990917 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 20.3 | 386.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 13.7 | 109.5 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 10.1 | 273.7 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 10.0 | 59.9 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 7.0 | 63.0 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) lipopolysaccharide receptor complex(GO:0046696) |
| 6.1 | 60.8 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 5.7 | 17.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 3.8 | 56.9 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 3.5 | 110.7 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 3.3 | 33.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 3.2 | 9.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 2.2 | 35.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 2.1 | 52.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 2.1 | 114.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 2.1 | 10.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 1.7 | 34.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 1.6 | 8.0 | GO:0033503 | HULC complex(GO:0033503) |
| 1.4 | 86.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 1.4 | 15.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 1.1 | 13.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 1.0 | 22.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.8 | 135.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.6 | 71.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.5 | 90.8 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.4 | 10.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.4 | 19.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.4 | 5.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 76.2 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.3 | 22.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.3 | 25.7 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.3 | 23.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 1.8 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.2 | 6.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 20.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 65.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.2 | 47.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 11.4 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 16.1 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 2.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 32.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 93.3 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 86.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 6.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 3.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 4.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 14.6 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 11.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 6.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 4.4 | GO:0045121 | membrane raft(GO:0045121) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 44.1 | 176.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 25.4 | 76.1 | GO:0002135 | CTP binding(GO:0002135) |
| 22.1 | 66.4 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 21.7 | 65.2 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 15.5 | 170.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 13.2 | 52.7 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 13.0 | 78.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 12.7 | 25.4 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 11.3 | 146.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 10.9 | 86.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 10.5 | 63.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 10.1 | 40.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 8.5 | 34.0 | GO:0070404 | NADH binding(GO:0070404) |
| 7.0 | 210.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 6.5 | 58.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 6.4 | 58.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 4.9 | 68.8 | GO:0046790 | virion binding(GO:0046790) |
| 3.8 | 132.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 3.4 | 17.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 3.3 | 20.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 3.2 | 159.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 3.2 | 9.5 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 3.0 | 21.0 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 2.7 | 13.7 | GO:1903135 | cupric ion binding(GO:1903135) |
| 2.7 | 32.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) N6-methyladenosine-containing RNA binding(GO:1990247) |
| 2.3 | 11.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 2.1 | 8.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 2.1 | 10.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 1.9 | 59.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 1.7 | 5.2 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 1.4 | 5.8 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 1.4 | 11.4 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 1.1 | 32.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 1.0 | 29.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 1.0 | 106.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.9 | 29.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.8 | 30.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.7 | 132.3 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.7 | 10.0 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.6 | 18.8 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.6 | 19.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.5 | 56.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.5 | 15.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.4 | 20.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.4 | 16.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.4 | 19.1 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.3 | 4.9 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.3 | 2.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.3 | 6.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 38.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.3 | 6.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 9.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 12.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 8.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 10.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 23.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 13.7 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 3.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 20.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 3.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 3.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 4.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 24.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.5 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 61.0 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 9.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 84.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 2.2 | 65.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 2.1 | 58.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 1.6 | 66.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 1.3 | 86.8 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 1.1 | 72.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 1.0 | 68.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 1.0 | 142.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.9 | 57.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.9 | 60.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.8 | 76.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.8 | 11.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.6 | 52.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.5 | 18.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.5 | 34.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.4 | 27.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.4 | 98.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.4 | 19.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.4 | 11.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 13.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 15.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 3.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 5.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 7.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.8 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.1 | 386.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 8.7 | 96.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 4.9 | 68.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 4.2 | 76.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 2.8 | 58.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 2.7 | 109.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 1.6 | 106.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 1.5 | 97.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 1.4 | 40.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 1.4 | 21.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 1.1 | 11.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 1.1 | 63.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.8 | 66.4 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.8 | 129.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.8 | 32.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.7 | 65.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.6 | 32.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.6 | 100.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.3 | 9.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 13.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 5.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 5.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 24.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 3.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 10.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 19.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 21.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 20.5 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 2.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |