Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for IRF6_IRF4_IRF5
Z-value: 0.33
Transcription factors associated with IRF6_IRF4_IRF5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
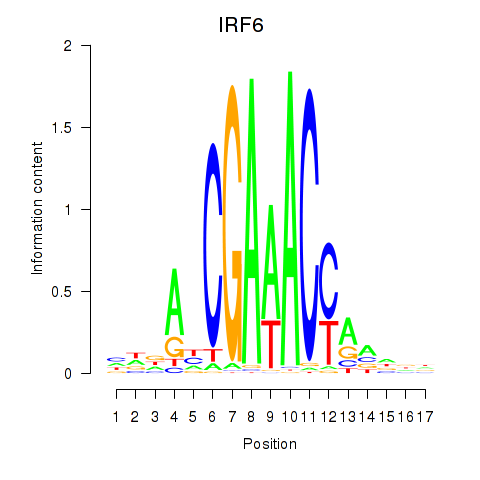
IRF6
|
ENSG00000117595.12 | IRF6 |
|
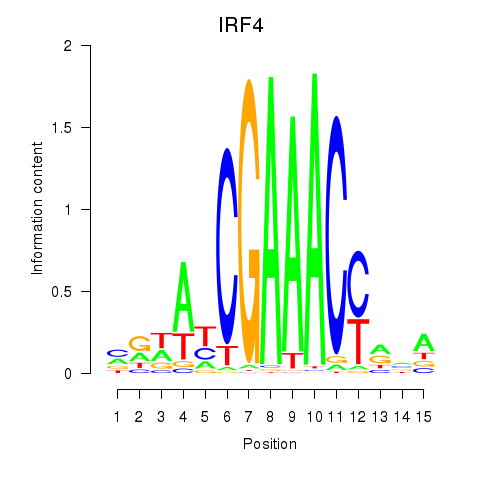
IRF4
|
ENSG00000137265.15 | IRF4 |
|
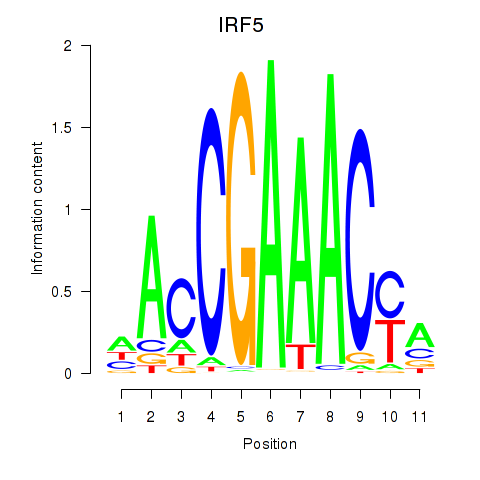
IRF5
|
ENSG00000128604.20 | IRF5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF6 | hg38_v1_chr1_-_209806124_209806175 | 0.16 | 1.5e-02 | Click! |
| IRF5 | hg38_v1_chr7_+_128937917_128937994 | 0.06 | 4.2e-01 | Click! |
| IRF4 | hg38_v1_chr6_+_391743_391759 | 0.03 | 6.5e-01 | Click! |
Activity profile of IRF6_IRF4_IRF5 motif
Sorted Z-values of IRF6_IRF4_IRF5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF6_IRF4_IRF5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.4 | 147.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 9.4 | 46.9 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 6.8 | 20.3 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 6.1 | 121.4 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 3.2 | 15.9 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 3.2 | 12.6 | GO:0032599 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 2.8 | 5.6 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 2.7 | 19.1 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 2.7 | 8.1 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 2.5 | 7.6 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 2.3 | 16.0 | GO:0052564 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 2.1 | 14.7 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 1.6 | 11.0 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 1.5 | 10.5 | GO:1905123 | regulation of glucosylceramidase activity(GO:1905123) |
| 1.4 | 7.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 1.3 | 3.9 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 1.0 | 12.0 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.9 | 7.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.9 | 12.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.8 | 2.5 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.8 | 2.4 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.7 | 9.0 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.7 | 2.8 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.7 | 4.8 | GO:1990668 | vesicle fusion with endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membrane(GO:1990668) |
| 0.7 | 6.0 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.7 | 5.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.6 | 7.1 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.6 | 6.8 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.5 | 2.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.5 | 1.6 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.5 | 17.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.5 | 7.8 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.5 | 1.9 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.4 | 1.3 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.4 | 5.9 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.4 | 19.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.4 | 2.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.4 | 2.0 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.4 | 13.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.4 | 3.0 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.4 | 1.5 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.3 | 21.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.3 | 6.7 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.3 | 3.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.3 | 2.0 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.3 | 0.8 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.3 | 3.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 1.0 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.3 | 5.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 3.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 2.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.2 | 12.1 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.2 | 6.0 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.2 | 0.4 | GO:1904907 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.2 | 0.7 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.2 | 4.2 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.2 | 2.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.2 | 1.5 | GO:0072564 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 3.2 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.2 | 1.8 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.2 | 3.5 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.2 | 7.1 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.2 | 7.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 5.4 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 4.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 3.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 2.1 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 7.3 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 3.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.6 | GO:0006064 | glucuronate catabolic process(GO:0006064) glucuronate catabolic process to xylulose 5-phosphate(GO:0019640) L-ascorbic acid biosynthetic process(GO:0019853) xylulose 5-phosphate metabolic process(GO:0051167) xylulose 5-phosphate biosynthetic process(GO:1901159) |
| 0.1 | 0.4 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 1.5 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 8.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 0.9 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 9.0 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 1.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 2.5 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 9.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 2.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 3.0 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 4.8 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 2.8 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 0.9 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 1.5 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 1.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 6.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.7 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 2.5 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 1.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 3.5 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.8 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 11.7 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 1.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 3.6 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.2 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.6 | GO:0009452 | 7-methylguanosine mRNA capping(GO:0006370) 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 | 0.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 1.4 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 3.6 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 1.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 12.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.3 | 6.7 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.0 | 4.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.9 | 5.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.6 | 2.4 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.5 | 5.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.5 | 12.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.5 | 3.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 2.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.4 | 0.8 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.4 | 3.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.4 | 18.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 22.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.3 | 6.9 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.3 | 7.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.3 | 4.8 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 9.1 | GO:0098553 | integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.3 | 6.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 3.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 36.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.3 | 145.5 | GO:0045121 | membrane raft(GO:0045121) |
| 0.3 | 10.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 36.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.3 | 2.0 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.3 | 2.0 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.3 | 112.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.2 | 10.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.2 | 1.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 12.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 2.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 3.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.4 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 8.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.5 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 1.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 11.8 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 2.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.3 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 22.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.9 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.9 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.9 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 7.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 3.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 14.7 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 29.7 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 11.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 2.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 3.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 2.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 6.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.4 | GO:0001726 | ruffle(GO:0001726) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.2 | 147.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 3.8 | 19.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 3.5 | 13.9 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 3.2 | 16.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 2.7 | 8.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 2.3 | 9.0 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 2.0 | 20.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 1.6 | 4.8 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 1.4 | 7.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 1.0 | 13.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.9 | 12.0 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.9 | 5.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.8 | 3.3 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.8 | 31.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.8 | 2.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.6 | 5.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.6 | 4.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.5 | 2.0 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.5 | 1.5 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.5 | 5.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.5 | 2.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.4 | 52.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.4 | 2.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 2.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 118.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.3 | 2.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 13.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.3 | 2.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.3 | 12.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.3 | 5.9 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.3 | 0.8 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 0.3 | 1.0 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.3 | 3.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 3.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 6.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 0.9 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 9.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 6.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 5.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 0.2 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.2 | 1.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 7.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 5.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 14.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 0.9 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.1 | 0.5 | GO:0005124 | N-formyl peptide receptor activity(GO:0004982) scavenger receptor binding(GO:0005124) |
| 0.1 | 2.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 1.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 2.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 3.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.9 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 28.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 9.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.3 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.1 | 1.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.6 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 5.0 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 1.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 43.2 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 0.8 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 4.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.8 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 5.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 6.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 6.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 11.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 1.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.7 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.0 | 1.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 3.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 6.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 4.2 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 1.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.5 | 19.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 6.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.3 | 13.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 46.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 7.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 11.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 14.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 2.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.2 | 32.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 7.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.2 | 3.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 6.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 6.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 6.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 3.9 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 4.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 4.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 2.0 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 3.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 2.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 46.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 1.5 | 157.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.7 | 5.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 13.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.4 | 20.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.3 | 5.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.3 | 9.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 6.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 24.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.3 | 5.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 5.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 7.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 18.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 5.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 3.7 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.2 | 3.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 3.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 5.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 5.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 5.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 14.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 3.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 12.8 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 2.4 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 1.0 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 3.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.9 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 0.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 3.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 3.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 5.2 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |