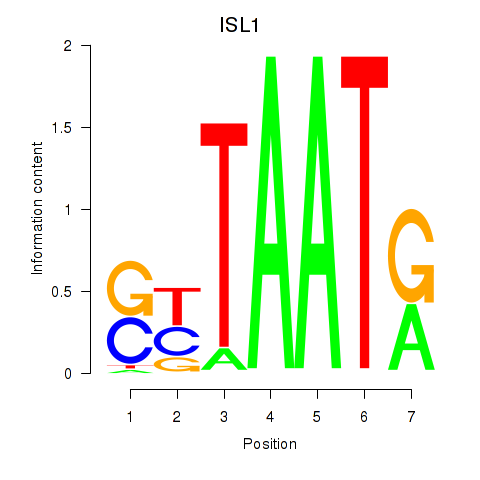
|
chr2_-_174846405
Show fit
|
63.53 |
ENST00000409597.5
ENST00000413882.6
|
CHN1
|
chimerin 1
|
|
chr4_-_176002332
Show fit
|
35.32 |
ENST00000280187.11
ENST00000512509.5
|
GPM6A
|
glycoprotein M6A
|
|
chr12_-_49897056
Show fit
|
26.57 |
ENST00000552863.5
|
FAIM2
|
Fas apoptotic inhibitory molecule 2
|
|
chr5_+_141370236
Show fit
|
26.48 |
ENST00000576222.2
ENST00000618934.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr14_-_59870752
Show fit
|
25.25 |
ENST00000611068.1
ENST00000267484.10
|
RTN1
|
reticulon 1
|
|
chr2_-_174847015
Show fit
|
24.82 |
ENST00000650938.1
|
CHN1
|
chimerin 1
|
|
chr1_-_156248084
Show fit
|
24.13 |
ENST00000652405.1
ENST00000335852.6
ENST00000540423.5
ENST00000612424.4
ENST00000613336.4
ENST00000623241.3
|
PAQR6
|
progestin and adipoQ receptor family member 6
|
|
chr6_-_52840843
Show fit
|
22.26 |
ENST00000370989.6
|
GSTA5
|
glutathione S-transferase alpha 5
|
|
chr1_-_156248013
Show fit
|
22.15 |
ENST00000368270.2
|
PAQR6
|
progestin and adipoQ receptor family member 6
|
|
chr18_-_24397784
Show fit
|
21.85 |
ENST00000399441.4
ENST00000319481.8
|
OSBPL1A
|
oxysterol binding protein like 1A
|
|
chr1_-_156248038
Show fit
|
20.81 |
ENST00000470198.5
ENST00000292291.10
ENST00000356983.7
|
PAQR6
|
progestin and adipoQ receptor family member 6
|
|
chr12_+_78864768
Show fit
|
19.65 |
ENST00000261205.9
ENST00000457153.6
|
SYT1
|
synaptotagmin 1
|
|
chr16_+_7332839
Show fit
|
18.65 |
ENST00000355637.9
|
RBFOX1
|
RNA binding fox-1 homolog 1
|
|
chr11_-_134412234
Show fit
|
18.58 |
ENST00000312527.9
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1
|
|
chr2_-_223602284
Show fit
|
18.58 |
ENST00000421386.1
ENST00000305409.3
ENST00000433889.1
|
SCG2
|
secretogranin II
|
|
chrX_+_55452119
Show fit
|
18.48 |
ENST00000342972.3
|
MAGEH1
|
MAGE family member H1
|
|
chr11_-_134411854
Show fit
|
18.19 |
ENST00000392580.5
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1
|
|
chr14_-_100569780
Show fit
|
17.85 |
ENST00000355173.7
|
BEGAIN
|
brain enriched guanylate kinase associated
|
|
chr5_+_140841183
Show fit
|
17.83 |
ENST00000378123.4
ENST00000531613.2
|
PCDHA8
|
protocadherin alpha 8
|
|
chr8_+_22053543
Show fit
|
17.62 |
ENST00000519850.5
ENST00000381470.7
|
DMTN
|
dematin actin binding protein
|
|
chr4_-_185956652
Show fit
|
17.28 |
ENST00000355634.9
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr12_+_78863962
Show fit
|
17.11 |
ENST00000393240.7
|
SYT1
|
synaptotagmin 1
|
|
chr6_-_116254063
Show fit
|
16.65 |
ENST00000420283.3
|
TSPYL4
|
TSPY like 4
|
|
chr2_+_172860038
Show fit
|
16.39 |
ENST00000538974.5
ENST00000540783.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4
|
|
chr3_+_35679614
Show fit
|
16.35 |
ENST00000474696.5
ENST00000412048.5
ENST00000396482.6
ENST00000432682.5
|
ARPP21
|
cAMP regulated phosphoprotein 21
|
|
chr18_-_5543988
Show fit
|
16.00 |
ENST00000341928.7
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3
|
|
chr15_+_80404320
Show fit
|
15.90 |
ENST00000303329.9
ENST00000622346.4
|
ARNT2
|
aryl hydrocarbon receptor nuclear translocator 2
|
|
chr2_+_95025700
Show fit
|
15.86 |
ENST00000309988.9
ENST00000353004.7
ENST00000354078.7
ENST00000349807.3
|
MAL
|
mal, T cell differentiation protein
|
|
chr12_-_91180365
Show fit
|
15.26 |
ENST00000547937.5
|
DCN
|
decorin
|
|
chr4_-_185956348
Show fit
|
15.25 |
ENST00000431902.5
ENST00000284776.11
ENST00000415274.5
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_+_174045673
Show fit
|
15.21 |
ENST00000303177.8
ENST00000519867.5
|
NSG2
|
neuronal vesicle trafficking associated 2
|
|
chr9_-_110999458
Show fit
|
14.46 |
ENST00000374430.6
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr18_-_5543960
Show fit
|
13.60 |
ENST00000400111.8
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3
|
|
chr3_+_159069252
Show fit
|
13.12 |
ENST00000640015.1
ENST00000476809.7
ENST00000485419.7
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough
|
|
chr18_-_5544250
Show fit
|
12.43 |
ENST00000540638.6
ENST00000342933.7
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3
|
|
chr4_+_113049479
Show fit
|
12.12 |
ENST00000671727.1
ENST00000671762.1
ENST00000672366.1
ENST00000672502.1
ENST00000672045.1
ENST00000672251.1
ENST00000672854.1
|
ANK2
|
ankyrin 2
|
|
chr5_-_138875290
Show fit
|
12.11 |
ENST00000521094.2
ENST00000274711.7
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr15_+_28885955
Show fit
|
12.07 |
ENST00000558402.5
ENST00000683413.1
ENST00000558330.5
|
APBA2
|
amyloid beta precursor protein binding family A member 2
|
|
chr17_-_55511434
Show fit
|
11.73 |
ENST00000636752.1
|
SMIM36
|
small integral membrane protein 36
|
|
chr4_-_86360039
Show fit
|
11.32 |
ENST00000515650.2
ENST00000641724.1
ENST00000641607.1
ENST00000641324.1
ENST00000641903.1
ENST00000395157.9
ENST00000641823.1
ENST00000641873.1
ENST00000641102.1
ENST00000641462.2
ENST00000641217.1
ENST00000642006.1
ENST00000641020.1
ENST00000641110.1
ENST00000639175.1
ENST00000641485.1
ENST00000641864.1
ENST00000641954.1
ENST00000641647.1
ENST00000641459.1
ENST00000641762.1
ENST00000641777.1
ENST00000641208.1
ENST00000642015.1
ENST00000641493.1
ENST00000642032.1
ENST00000641010.1
ENST00000641287.1
ENST00000641943.1
ENST00000642103.1
ENST00000641047.1
ENST00000641166.1
ENST00000641207.1
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr4_-_86360010
Show fit
|
11.25 |
ENST00000641911.1
ENST00000641072.1
ENST00000359221.8
ENST00000640490.1
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr22_+_39994926
Show fit
|
10.30 |
ENST00000333407.11
|
FAM83F
|
family with sequence similarity 83 member F
|
|
chr4_+_113049616
Show fit
|
10.23 |
ENST00000504454.5
ENST00000357077.9
ENST00000394537.7
ENST00000672779.1
ENST00000264366.10
|
ANK2
|
ankyrin 2
|
|
chrX_+_136197039
Show fit
|
9.94 |
ENST00000370683.6
|
FHL1
|
four and a half LIM domains 1
|
|
chr5_+_140868945
Show fit
|
9.83 |
ENST00000398640.7
|
PCDHA11
|
protocadherin alpha 11
|
|
chr4_-_86360071
Show fit
|
9.53 |
ENST00000641677.1
ENST00000639234.1
ENST00000641553.1
ENST00000641826.1
ENST00000641537.1
ENST00000395169.9
ENST00000641408.1
ENST00000638225.1
ENST00000641052.1
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr1_-_94925759
Show fit
|
9.43 |
ENST00000415017.1
ENST00000545882.5
|
CNN3
|
calponin 3
|
|
chr11_-_35360050
Show fit
|
9.30 |
ENST00000644868.1
ENST00000643454.1
ENST00000646080.1
|
SLC1A2
|
solute carrier family 1 member 2
|
|
chr5_+_140875299
Show fit
|
8.90 |
ENST00000613593.1
ENST00000398631.3
|
PCDHA12
|
protocadherin alpha 12
|
|
chr2_+_172928165
Show fit
|
8.48 |
ENST00000535187.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4
|
|
chr18_-_55510753
Show fit
|
8.27 |
ENST00000543082.5
|
TCF4
|
transcription factor 4
|
|
chr9_+_101185029
Show fit
|
7.97 |
ENST00000395056.2
|
PLPPR1
|
phospholipid phosphatase related 1
|
|
chr1_-_113887574
Show fit
|
7.96 |
ENST00000393316.8
|
BCL2L15
|
BCL2 like 15
|
|
chr4_+_61201223
Show fit
|
7.56 |
ENST00000512091.6
|
ADGRL3
|
adhesion G protein-coupled receptor L3
|
|
chr1_+_248231417
Show fit
|
7.42 |
ENST00000641868.1
|
OR2M4
|
olfactory receptor family 2 subfamily M member 4
|
|
chr8_-_19682576
Show fit
|
7.35 |
ENST00000332246.10
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1
|
|
chr8_-_100559702
Show fit
|
7.35 |
ENST00000520311.5
ENST00000520552.5
ENST00000521345.1
ENST00000523000.5
ENST00000335659.7
ENST00000358990.3
ENST00000519597.5
|
ANKRD46
|
ankyrin repeat domain 46
|
|
chrX_+_136169624
Show fit
|
7.28 |
ENST00000394153.6
|
FHL1
|
four and a half LIM domains 1
|
|
chrX_-_103686687
Show fit
|
7.20 |
ENST00000441076.7
ENST00000422355.5
ENST00000442614.5
ENST00000451301.5
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr9_-_14300231
Show fit
|
7.18 |
ENST00000636735.1
|
NFIB
|
nuclear factor I B
|
|
chr10_-_60572599
Show fit
|
7.12 |
ENST00000503366.5
|
ANK3
|
ankyrin 3
|
|
chr11_-_70717994
Show fit
|
7.05 |
ENST00000659264.1
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr5_+_141397942
Show fit
|
6.97 |
ENST00000617380.2
ENST00000621169.1
|
PCDHGB5
|
protocadherin gamma subfamily B, 5
|
|
chrX_+_136169833
Show fit
|
6.88 |
ENST00000628032.2
|
FHL1
|
four and a half LIM domains 1
|
|
chr2_-_166149120
Show fit
|
6.14 |
ENST00000641575.1
ENST00000641603.1
|
SCN1A
|
sodium voltage-gated channel alpha subunit 1
|
|
chr6_+_36885944
Show fit
|
6.11 |
ENST00000480824.7
|
C6orf89
|
chromosome 6 open reading frame 89
|
|
chr18_-_55423757
Show fit
|
6.09 |
ENST00000675707.1
|
TCF4
|
transcription factor 4
|
|
chr2_-_166149204
Show fit
|
6.00 |
ENST00000635750.1
|
SCN1A
|
sodium voltage-gated channel alpha subunit 1
|
|
chr8_+_38728186
Show fit
|
5.87 |
ENST00000519416.5
ENST00000520615.5
|
TACC1
|
transforming acidic coiled-coil containing protein 1
|
|
chrX_+_136169891
Show fit
|
5.65 |
ENST00000449474.5
|
FHL1
|
four and a half LIM domains 1
|
|
chr16_+_47462112
Show fit
|
5.47 |
ENST00000567200.1
|
PHKB
|
phosphorylase kinase regulatory subunit beta
|
|
chrX_-_72306962
Show fit
|
5.25 |
ENST00000246139.9
|
CITED1
|
Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 1
|
|
chr6_+_150683593
Show fit
|
4.98 |
ENST00000644968.1
|
PLEKHG1
|
pleckstrin homology and RhoGEF domain containing G1
|
|
chr19_-_17377334
Show fit
|
4.76 |
ENST00000252590.9
ENST00000599426.1
|
PLVAP
|
plasmalemma vesicle associated protein
|
|
chr5_-_95081482
Show fit
|
4.73 |
ENST00000312216.12
ENST00000512425.5
ENST00000505208.5
ENST00000429576.6
ENST00000508509.5
ENST00000510732.5
|
MCTP1
|
multiple C2 and transmembrane domain containing 1
|
|
chrX_-_72307148
Show fit
|
4.69 |
ENST00000453707.6
ENST00000373619.7
|
CITED1
|
Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 1
|
|
chr18_+_48539112
Show fit
|
4.68 |
ENST00000382998.8
|
CTIF
|
cap binding complex dependent translation initiation factor
|
|
chr14_+_19743571
Show fit
|
4.66 |
ENST00000642117.2
|
OR4Q3
|
olfactory receptor family 4 subfamily Q member 3
|
|
chr15_+_42359577
Show fit
|
4.62 |
ENST00000357568.8
|
CAPN3
|
calpain 3
|
|
chr12_+_80716906
Show fit
|
4.59 |
ENST00000228644.4
|
MYF5
|
myogenic factor 5
|
|
chr18_+_48539017
Show fit
|
4.57 |
ENST00000256413.8
|
CTIF
|
cap binding complex dependent translation initiation factor
|
|
chr15_+_42359454
Show fit
|
4.48 |
ENST00000349748.8
ENST00000318023.11
ENST00000397163.8
|
CAPN3
|
calpain 3
|
|
chr6_-_52763473
Show fit
|
4.44 |
ENST00000493422.3
|
GSTA2
|
glutathione S-transferase alpha 2
|
|
chrY_-_6872608
Show fit
|
4.30 |
ENST00000383036.1
|
AMELY
|
amelogenin Y-linked
|
|
chr16_+_2148603
Show fit
|
4.19 |
ENST00000210187.11
|
RAB26
|
RAB26, member RAS oncogene family
|
|
chr13_-_108218293
Show fit
|
4.17 |
ENST00000405925.2
|
LIG4
|
DNA ligase 4
|
|
chr1_-_79006773
Show fit
|
4.17 |
ENST00000671209.1
|
ADGRL4
|
adhesion G protein-coupled receptor L4
|
|
chr2_+_233307806
Show fit
|
4.15 |
ENST00000447536.5
ENST00000409110.6
|
SAG
|
S-antigen visual arrestin
|
|
chr1_+_100133135
Show fit
|
4.15 |
ENST00000370143.5
ENST00000370141.7
|
TRMT13
|
tRNA methyltransferase 13 homolog
|
|
chr19_-_49119092
Show fit
|
4.07 |
ENST00000408991.4
|
C19orf73
|
chromosome 19 open reading frame 73
|
|
chr6_+_31547560
Show fit
|
4.06 |
ENST00000376148.9
ENST00000376145.8
|
NFKBIL1
|
NFKB inhibitor like 1
|
|
chr1_+_196652022
Show fit
|
3.98 |
ENST00000367429.9
ENST00000630130.2
ENST00000359637.2
|
CFH
|
complement factor H
|
|
chr3_-_192727500
Show fit
|
3.89 |
ENST00000430714.5
ENST00000418610.1
ENST00000445105.7
ENST00000684728.1
ENST00000448795.5
ENST00000683935.1
ENST00000684282.1
ENST00000683451.1
|
FGF12
|
fibroblast growth factor 12
|
|
chr12_+_119668109
Show fit
|
3.80 |
ENST00000229328.10
ENST00000630317.1
|
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1
|
|
chrX_+_136169664
Show fit
|
3.76 |
ENST00000456445.5
|
FHL1
|
four and a half LIM domains 1
|
|
chr12_-_42237727
Show fit
|
3.75 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2
|
|
chr7_+_90709530
Show fit
|
3.74 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14
|
|
chr3_+_4680617
Show fit
|
3.60 |
ENST00000648212.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1
|
|
chr16_+_55656294
Show fit
|
3.53 |
ENST00000379906.6
|
SLC6A2
|
solute carrier family 6 member 2
|
|
chr1_-_79006680
Show fit
|
3.48 |
ENST00000370742.4
ENST00000656841.1
|
ADGRL4
|
adhesion G protein-coupled receptor L4
|
|
chr2_-_54987578
Show fit
|
3.44 |
ENST00000486085.5
|
RTN4
|
reticulon 4
|
|
chr11_-_63015804
Show fit
|
3.43 |
ENST00000535878.5
ENST00000545207.5
|
SLC22A8
|
solute carrier family 22 member 8
|
|
chr21_-_46155567
Show fit
|
3.31 |
ENST00000291670.9
ENST00000397748.5
ENST00000397743.1
ENST00000397746.8
|
FTCD
|
formimidoyltransferase cyclodeaminase
|
|
chr6_+_43014103
Show fit
|
3.30 |
ENST00000244670.12
ENST00000326974.9
ENST00000332245.9
|
KLHDC3
|
kelch domain containing 3
|
|
chr18_+_74499861
Show fit
|
3.14 |
ENST00000324301.12
|
CNDP2
|
carnosine dipeptidase 2
|
|
chr2_+_170178136
Show fit
|
3.11 |
ENST00000409044.7
ENST00000408978.9
|
MYO3B
|
myosin IIIB
|
|
chr4_+_56436233
Show fit
|
3.10 |
ENST00000512576.3
|
PAICS
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase
|
|
chr7_+_90709231
Show fit
|
3.08 |
ENST00000446790.5
ENST00000265741.7
|
CDK14
|
cyclin dependent kinase 14
|
|
chrX_+_51893533
Show fit
|
3.03 |
ENST00000375722.5
ENST00000326587.12
ENST00000375695.2
|
MAGED1
|
MAGE family member D1
|
|
chr10_-_77637789
Show fit
|
2.99 |
ENST00000481070.1
ENST00000640969.1
ENST00000286628.14
ENST00000638991.1
ENST00000639913.1
ENST00000480683.2
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr11_-_63015831
Show fit
|
2.98 |
ENST00000430500.6
ENST00000336232.7
|
SLC22A8
|
solute carrier family 22 member 8
|
|
chr9_+_37667997
Show fit
|
2.97 |
ENST00000539465.5
|
FRMPD1
|
FERM and PDZ domain containing 1
|
|
chr12_-_10098977
Show fit
|
2.95 |
ENST00000315330.8
ENST00000457018.6
|
CLEC1A
|
C-type lectin domain family 1 member A
|
|
chr19_-_18606779
Show fit
|
2.93 |
ENST00000684169.1
ENST00000392386.8
|
CRLF1
|
cytokine receptor like factor 1
|
|
chr4_+_87832917
Show fit
|
2.86 |
ENST00000395102.8
ENST00000497649.6
ENST00000540395.1
ENST00000560249.5
ENST00000511670.5
ENST00000361056.3
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chr16_+_55656089
Show fit
|
2.85 |
ENST00000414754.7
|
SLC6A2
|
solute carrier family 6 member 2
|
|
chr1_-_160285120
Show fit
|
2.84 |
ENST00000368072.10
|
PEX19
|
peroxisomal biogenesis factor 19
|
|
chr8_-_12194067
Show fit
|
2.82 |
ENST00000524571.6
ENST00000533852.6
ENST00000533513.1
ENST00000448228.6
ENST00000534520.5
|
FAM86B1
|
family with sequence similarity 86 member B1
|
|
chr19_+_3506251
Show fit
|
2.81 |
ENST00000441788.7
|
FZR1
|
fizzy and cell division cycle 20 related 1
|
|
chr6_-_100464912
Show fit
|
2.79 |
ENST00000369208.8
|
SIM1
|
SIM bHLH transcription factor 1
|
|
chr21_-_40847149
Show fit
|
2.68 |
ENST00000400454.6
|
DSCAM
|
DS cell adhesion molecule
|
|
chr5_-_16508858
Show fit
|
2.68 |
ENST00000684456.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr6_-_136466858
Show fit
|
2.68 |
ENST00000544465.5
|
MAP7
|
microtubule associated protein 7
|
|
chr5_+_32710630
Show fit
|
2.63 |
ENST00000326958.5
|
NPR3
|
natriuretic peptide receptor 3
|
|
chr7_-_108240049
Show fit
|
2.54 |
ENST00000379022.8
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr10_+_122163672
Show fit
|
2.47 |
ENST00000369004.7
ENST00000260733.7
|
TACC2
|
transforming acidic coiled-coil containing protein 2
|
|
chr17_+_28371656
Show fit
|
2.44 |
ENST00000585482.6
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr7_-_78771265
Show fit
|
2.40 |
ENST00000630991.2
ENST00000629359.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr6_+_27388748
Show fit
|
2.31 |
ENST00000244576.9
|
ZNF391
|
zinc finger protein 391
|
|
chr11_+_22666604
Show fit
|
2.29 |
ENST00000454584.6
|
GAS2
|
growth arrest specific 2
|
|
chr16_+_55656249
Show fit
|
2.23 |
ENST00000219833.13
ENST00000574918.2
|
SLC6A2
|
solute carrier family 6 member 2
|
|
chr9_+_27109200
Show fit
|
2.23 |
ENST00000380036.10
|
TEK
|
TEK receptor tyrosine kinase
|
|
chrX_-_23907887
Show fit
|
2.16 |
ENST00000379226.9
|
APOO
|
apolipoprotein O
|
|
chr17_-_15598618
Show fit
|
2.11 |
ENST00000583965.5
|
CDRT1
|
CMT1A duplicated region transcript 1
|
|
chr7_-_44541318
Show fit
|
2.07 |
ENST00000381160.8
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1
|
|
chr16_+_55655960
Show fit
|
2.00 |
ENST00000568943.6
|
SLC6A2
|
solute carrier family 6 member 2
|
|
chr9_+_27109135
Show fit
|
1.93 |
ENST00000519097.5
ENST00000615002.4
|
TEK
|
TEK receptor tyrosine kinase
|
|
chr8_+_109362453
Show fit
|
1.93 |
ENST00000378402.10
|
PKHD1L1
|
PKHD1 like 1
|
|
chr5_+_175861628
Show fit
|
1.87 |
ENST00000509837.5
|
CPLX2
|
complexin 2
|
|
chr10_+_133527355
Show fit
|
1.79 |
ENST00000252945.8
ENST00000421586.5
ENST00000418356.1
|
CYP2E1
|
cytochrome P450 family 2 subfamily E member 1
|
|
chr11_-_75096876
Show fit
|
1.79 |
ENST00000641541.1
ENST00000641593.1
ENST00000641504.1
ENST00000641931.1
ENST00000647690.1
|
OR2AT4
ENSG00000284722.2
|
olfactory receptor family 2 subfamily AT member 4
novel transcript
|
|
chrX_-_30309387
Show fit
|
1.77 |
ENST00000378970.5
|
NR0B1
|
nuclear receptor subfamily 0 group B member 1
|
|
chr2_-_207165923
Show fit
|
1.72 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7
|
|
chr1_-_7940825
Show fit
|
1.72 |
ENST00000377507.8
|
TNFRSF9
|
TNF receptor superfamily member 9
|
|
chr10_-_77637902
Show fit
|
1.63 |
ENST00000286627.10
ENST00000639486.1
ENST00000640523.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr7_-_44541262
Show fit
|
1.62 |
ENST00000289547.8
ENST00000546276.5
ENST00000423141.1
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1
|
|
chr6_+_168017873
Show fit
|
1.60 |
ENST00000351261.4
ENST00000354419.6
|
KIF25
|
kinesin family member 25
|
|
chr6_-_110815408
Show fit
|
1.58 |
ENST00000368911.8
|
CDK19
|
cyclin dependent kinase 19
|
|
chr12_-_30735014
Show fit
|
1.54 |
ENST00000433722.6
|
CAPRIN2
|
caprin family member 2
|
|
chr5_+_137867852
Show fit
|
1.49 |
ENST00000421631.6
ENST00000239926.9
|
MYOT
|
myotilin
|
|
chr7_+_144070313
Show fit
|
1.40 |
ENST00000641441.1
|
OR2A25
|
olfactory receptor family 2 subfamily A member 25
|
|
chr12_+_54008961
Show fit
|
1.40 |
ENST00000040584.6
|
HOXC8
|
homeobox C8
|
|
chr3_+_38265802
Show fit
|
1.34 |
ENST00000311856.9
|
SLC22A13
|
solute carrier family 22 member 13
|
|
chr11_+_57805541
Show fit
|
1.16 |
ENST00000683201.1
ENST00000683769.1
|
CTNND1
|
catenin delta 1
|
|
chr3_-_119557728
Show fit
|
1.14 |
ENST00000383669.3
|
CD80
|
CD80 molecule
|
|
chr13_-_99016034
Show fit
|
1.12 |
ENST00000448493.7
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr15_+_58987652
Show fit
|
1.00 |
ENST00000348370.9
ENST00000559160.1
|
RNF111
|
ring finger protein 111
|
|
chr3_+_119782094
Show fit
|
0.95 |
ENST00000393716.8
|
NR1I2
|
nuclear receptor subfamily 1 group I member 2
|
|
chr1_+_197413827
Show fit
|
0.90 |
ENST00000367397.1
ENST00000681519.1
|
CRB1
|
crumbs cell polarity complex component 1
|
|
chr4_+_70195719
Show fit
|
0.84 |
ENST00000683306.1
|
ODAM
|
odontogenic, ameloblast associated
|
|
chr8_-_78805306
Show fit
|
0.84 |
ENST00000639719.1
ENST00000263851.9
|
IL7
|
interleukin 7
|
|
chr20_+_43507668
Show fit
|
0.83 |
ENST00000434666.5
ENST00000649084.1
ENST00000418998.7
ENST00000427442.8
ENST00000649917.1
|
L3MBTL1
|
L3MBTL histone methyl-lysine binding protein 1
|
|
chr17_-_66229380
Show fit
|
0.77 |
ENST00000205948.11
|
APOH
|
apolipoprotein H
|
|
chr17_-_48968587
Show fit
|
0.75 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide
|
|
chr5_+_141213919
Show fit
|
0.62 |
ENST00000341948.6
|
PCDHB13
|
protocadherin beta 13
|
|
chr11_-_18236795
Show fit
|
0.60 |
ENST00000278222.7
|
SAA4
|
serum amyloid A4, constitutive
|
|
chrX_-_100874351
Show fit
|
0.59 |
ENST00000372966.8
|
NOX1
|
NADPH oxidase 1
|
|
chr3_-_124887353
Show fit
|
0.54 |
ENST00000296181.9
|
ITGB5
|
integrin subunit beta 5
|
|
chr9_-_92404559
Show fit
|
0.54 |
ENST00000262551.8
ENST00000375561.10
|
OGN
|
osteoglycin
|
|
chr2_-_96740034
Show fit
|
0.53 |
ENST00000264963.9
ENST00000377079.8
|
LMAN2L
|
lectin, mannose binding 2 like
|
|
chr6_+_31739948
Show fit
|
0.51 |
ENST00000375755.8
ENST00000425703.5
ENST00000375750.9
ENST00000375703.7
ENST00000375740.7
|
MSH5
|
mutS homolog 5
|
|
chr16_+_20764036
Show fit
|
0.49 |
ENST00000440284.6
|
ACSM3
|
acyl-CoA synthetase medium chain family member 3
|
|
chr18_-_13915531
Show fit
|
0.47 |
ENST00000327606.4
|
MC2R
|
melanocortin 2 receptor
|
|
chrX_-_100874209
Show fit
|
0.45 |
ENST00000372964.5
ENST00000217885.5
|
NOX1
|
NADPH oxidase 1
|
|
chr19_+_3506355
Show fit
|
0.41 |
ENST00000652521.1
|
FZR1
|
fizzy and cell division cycle 20 related 1
|
|
chr16_-_23453136
Show fit
|
0.38 |
ENST00000307149.10
|
COG7
|
component of oligomeric golgi complex 7
|
|
chr9_+_133459965
Show fit
|
0.37 |
ENST00000540581.5
ENST00000542192.5
ENST00000291722.11
ENST00000316948.9
|
CACFD1
|
calcium channel flower domain containing 1
|
|
chr2_+_28395511
Show fit
|
0.36 |
ENST00000436647.1
|
FOSL2
|
FOS like 2, AP-1 transcription factor subunit
|
|
chr1_+_81800368
Show fit
|
0.29 |
ENST00000674489.1
ENST00000674442.1
ENST00000674419.1
ENST00000674407.1
ENST00000674168.1
ENST00000674307.1
ENST00000674209.1
ENST00000370715.5
ENST00000370713.5
ENST00000319517.10
ENST00000627151.2
ENST00000370717.6
|
ADGRL2
|
adhesion G protein-coupled receptor L2
|
|
chr8_-_132625378
Show fit
|
0.26 |
ENST00000522789.5
|
LRRC6
|
leucine rich repeat containing 6
|
|
chr17_-_69244846
Show fit
|
0.21 |
ENST00000269081.8
|
ABCA10
|
ATP binding cassette subfamily A member 10
|
|
chr4_-_99435396
Show fit
|
0.18 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
chr10_+_49610297
Show fit
|
0.17 |
ENST00000374115.5
|
SLC18A3
|
solute carrier family 18 member A3
|
|
chr7_+_130293134
Show fit
|
0.10 |
ENST00000445470.6
ENST00000492072.5
ENST00000222482.10
ENST00000473956.5
ENST00000493259.5
ENST00000486598.1
|
CPA4
|
carboxypeptidase A4
|
|
chr11_-_68004043
Show fit
|
0.10 |
ENST00000227471.7
|
UNC93B1
|
unc-93 homolog B1, TLR signaling regulator
|
|
chr2_+_225399684
Show fit
|
0.09 |
ENST00000636099.1
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2
|
|
chr14_+_21990357
Show fit
|
0.04 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16
|
|
chr12_-_52367478
Show fit
|
0.03 |
ENST00000257901.7
|
KRT85
|
keratin 85
|