Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for LHX3
Z-value: 0.82
Transcription factors associated with LHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX3
|
ENSG00000107187.17 | LHX3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX3 | hg38_v1_chr9_-_136203183_136203194, hg38_v1_chr9_-_136205122_136205133 | -0.36 | 6.3e-08 | Click! |
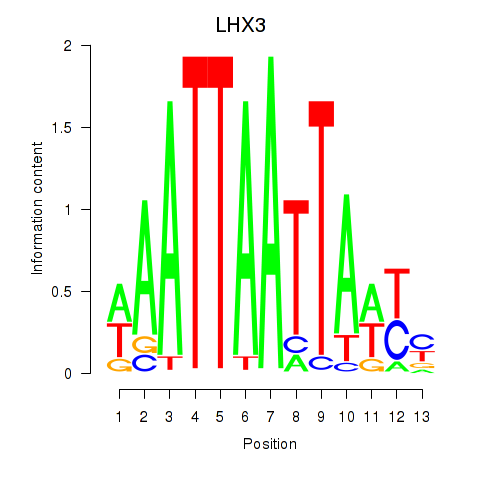
Activity profile of LHX3 motif
Sorted Z-values of LHX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_26103922 | 6.73 |
ENST00000377803.4
|
H4C3
|
H4 clustered histone 3 |
| chr14_+_34993240 | 6.10 |
ENST00000677647.1
|
SRP54
|
signal recognition particle 54 |
| chr1_-_150235972 | 5.58 |
ENST00000534220.1
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr6_+_106360668 | 5.29 |
ENST00000633556.3
|
CRYBG1
|
crystallin beta-gamma domain containing 1 |
| chr1_-_23799533 | 5.03 |
ENST00000429356.5
|
GALE
|
UDP-galactose-4-epimerase |
| chr3_-_64019334 | 4.35 |
ENST00000480205.5
|
PSMD6
|
proteasome 26S subunit, non-ATPase 6 |
| chr13_+_30422487 | 4.33 |
ENST00000638137.1
ENST00000635918.2 |
UBE2L5
|
ubiquitin conjugating enzyme E2 L5 |
| chr1_-_150236064 | 4.12 |
ENST00000532744.2
ENST00000369114.9 ENST00000369115.3 ENST00000583931.6 |
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr1_-_150235995 | 4.07 |
ENST00000436748.6
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr1_-_150235943 | 3.54 |
ENST00000533654.5
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr3_-_142029108 | 3.43 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr4_+_112647059 | 3.20 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein 7, transcriptional regulator |
| chr7_-_36724457 | 3.17 |
ENST00000617537.5
ENST00000435386.1 |
AOAH
|
acyloxyacyl hydrolase |
| chr12_-_118190510 | 3.11 |
ENST00000540561.5
ENST00000537952.1 ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr16_-_28623560 | 3.07 |
ENST00000350842.8
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chrX_+_108045050 | 3.07 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chrX_+_108044967 | 3.03 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr16_+_67570741 | 2.99 |
ENST00000644753.1
ENST00000642819.1 ENST00000645306.1 |
CTCF
|
CCCTC-binding factor |
| chr7_-_36724543 | 2.98 |
ENST00000612871.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr9_+_69145463 | 2.89 |
ENST00000636438.1
|
TJP2
|
tight junction protein 2 |
| chr15_+_64387828 | 2.48 |
ENST00000261884.8
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr14_+_61697622 | 2.43 |
ENST00000539097.2
|
HIF1A
|
hypoxia inducible factor 1 subunit alpha |
| chr12_-_10130241 | 2.39 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr15_+_65550819 | 2.38 |
ENST00000569894.5
|
HACD3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr12_-_7503841 | 2.35 |
ENST00000359156.8
|
CD163
|
CD163 molecule |
| chr9_-_125484490 | 2.31 |
ENST00000444226.1
|
MAPKAP1
|
MAPK associated protein 1 |
| chr17_-_40937641 | 2.24 |
ENST00000209718.8
|
KRT23
|
keratin 23 |
| chr12_-_10130143 | 2.21 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr18_+_58341038 | 2.18 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr1_-_45522870 | 2.13 |
ENST00000424390.2
|
PRDX1
|
peroxiredoxin 1 |
| chrX_-_13817027 | 2.11 |
ENST00000493677.5
ENST00000355135.6 ENST00000316715.9 |
GPM6B
|
glycoprotein M6B |
| chr17_-_40937445 | 1.99 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr11_-_63608542 | 1.95 |
ENST00000540943.1
|
PLAAT3
|
phospholipase A and acyltransferase 3 |
| chr10_+_92591733 | 1.95 |
ENST00000676647.1
|
KIF11
|
kinesin family member 11 |
| chr12_-_7503744 | 1.94 |
ENST00000396620.7
ENST00000432237.3 |
CD163
|
CD163 molecule |
| chr16_+_15395745 | 1.86 |
ENST00000287594.7
ENST00000396385.4 ENST00000568766.1 |
MPV17L
ENSG00000261130.5
|
MPV17 mitochondrial inner membrane protein like novel protein |
| chr4_+_41935114 | 1.83 |
ENST00000508448.5
ENST00000513702.5 ENST00000325094.9 |
TMEM33
|
transmembrane protein 33 |
| chr19_+_49513353 | 1.68 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr1_+_207325629 | 1.61 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr19_+_49513154 | 1.58 |
ENST00000426395.7
ENST00000600273.5 ENST00000599988.5 |
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr17_+_37491464 | 1.53 |
ENST00000613659.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr3_+_138621225 | 1.48 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr3_-_47475811 | 1.46 |
ENST00000265565.10
ENST00000428413.5 |
SCAP
|
SREBF chaperone |
| chr9_+_122370523 | 1.44 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr5_+_68290637 | 1.39 |
ENST00000336483.9
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr14_+_39114289 | 1.32 |
ENST00000396249.7
ENST00000250379.13 ENST00000534684.7 ENST00000308317.12 ENST00000527381.2 |
GEMIN2
|
gem nuclear organelle associated protein 2 |
| chr2_-_98663464 | 1.29 |
ENST00000414521.6
|
MGAT4A
|
alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A |
| chr7_+_92057602 | 1.24 |
ENST00000491695.2
|
AKAP9
|
A-kinase anchoring protein 9 |
| chr3_+_158110052 | 1.15 |
ENST00000295930.7
ENST00000471994.5 ENST00000482822.3 ENST00000476899.6 ENST00000683899.1 ENST00000684604.1 ENST00000682164.1 ENST00000464171.5 ENST00000611884.5 ENST00000312179.10 ENST00000475278.6 |
RSRC1
|
arginine and serine rich coiled-coil 1 |
| chr3_+_141387801 | 1.14 |
ENST00000514251.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr9_+_122371036 | 1.13 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr3_+_138621207 | 1.12 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr3_+_69763726 | 1.06 |
ENST00000448226.9
|
MITF
|
melanocyte inducing transcription factor |
| chr9_-_72953047 | 1.05 |
ENST00000297785.8
ENST00000376939.5 |
ALDH1A1
|
aldehyde dehydrogenase 1 family member A1 |
| chrX_-_107000185 | 1.02 |
ENST00000355610.9
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr1_-_158426237 | 0.94 |
ENST00000641042.1
|
OR10K2
|
olfactory receptor family 10 subfamily K member 2 |
| chr3_+_63967738 | 0.94 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr5_-_88823763 | 0.85 |
ENST00000635898.1
ENST00000626391.2 ENST00000628656.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr8_-_18083278 | 0.84 |
ENST00000636691.1
|
ASAH1
|
N-acylsphingosine amidohydrolase 1 |
| chr6_+_82363199 | 0.83 |
ENST00000535040.4
|
TPBG
|
trophoblast glycoprotein |
| chr4_+_41613476 | 0.81 |
ENST00000508466.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr9_+_122371014 | 0.78 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr3_+_41194741 | 0.69 |
ENST00000643541.1
ENST00000426215.5 ENST00000645210.1 ENST00000646381.1 ENST00000405570.6 ENST00000642248.1 ENST00000433400.6 |
CTNNB1
|
catenin beta 1 |
| chr17_-_44066595 | 0.67 |
ENST00000585388.2
ENST00000293406.8 |
LSM12
|
LSM12 homolog |
| chr7_+_107583919 | 0.63 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr11_+_60056587 | 0.62 |
ENST00000395032.6
ENST00000358152.6 |
MS4A3
|
membrane spanning 4-domains A3 |
| chr18_-_24311495 | 0.61 |
ENST00000357041.8
|
OSBPL1A
|
oxysterol binding protein like 1A |
| chr18_-_24272179 | 0.59 |
ENST00000399443.7
|
OSBPL1A
|
oxysterol binding protein like 1A |
| chr3_-_12545499 | 0.54 |
ENST00000564146.4
|
MKRN2OS
|
MKRN2 opposite strand |
| chr15_-_19988117 | 0.53 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr12_-_42237727 | 0.52 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr8_-_79767462 | 0.52 |
ENST00000674295.1
ENST00000518733.1 ENST00000674418.1 ENST00000674358.1 ENST00000354724.8 |
HEY1
|
hes related family bHLH transcription factor with YRPW motif 1 |
| chr1_+_244051275 | 0.51 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr5_-_79514127 | 0.50 |
ENST00000334082.11
|
HOMER1
|
homer scaffold protein 1 |
| chr19_+_14583076 | 0.50 |
ENST00000547437.5
ENST00000417570.6 |
CLEC17A
|
C-type lectin domain containing 17A |
| chr5_-_39274515 | 0.50 |
ENST00000510188.1
|
FYB1
|
FYN binding protein 1 |
| chr10_-_114144599 | 0.49 |
ENST00000428953.1
|
CCDC186
|
coiled-coil domain containing 186 |
| chr19_+_9185594 | 0.48 |
ENST00000344248.4
|
OR7D2
|
olfactory receptor family 7 subfamily D member 2 |
| chr4_-_119322128 | 0.46 |
ENST00000274024.4
|
FABP2
|
fatty acid binding protein 2 |
| chr7_+_138460238 | 0.46 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr11_+_60056653 | 0.45 |
ENST00000278865.8
|
MS4A3
|
membrane spanning 4-domains A3 |
| chr7_-_123199960 | 0.44 |
ENST00000194130.7
|
SLC13A1
|
solute carrier family 13 member 1 |
| chr20_+_45416551 | 0.41 |
ENST00000639292.1
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis class T |
| chr18_+_74534493 | 0.38 |
ENST00000358821.8
|
CNDP1
|
carnosine dipeptidase 1 |
| chr6_+_113857333 | 0.38 |
ENST00000612661.2
|
MARCKS
|
myristoylated alanine rich protein kinase C substrate |
| chr4_+_53377630 | 0.36 |
ENST00000337488.11
ENST00000358575.9 ENST00000507922.5 ENST00000306932.10 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr2_-_2324642 | 0.33 |
ENST00000650485.1
ENST00000649207.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr3_-_165078480 | 0.32 |
ENST00000264382.8
|
SI
|
sucrase-isomaltase |
| chr16_-_28623330 | 0.31 |
ENST00000677940.1
|
ENSG00000288656.1
|
novel protein |
| chr19_-_48646155 | 0.31 |
ENST00000084798.9
|
CA11
|
carbonic anhydrase 11 |
| chr14_+_22494810 | 0.30 |
ENST00000390493.1
|
TRAJ44
|
T cell receptor alpha joining 44 |
| chrX_+_109535775 | 0.26 |
ENST00000218004.5
|
NXT2
|
nuclear transport factor 2 like export factor 2 |
| chr13_-_60013178 | 0.25 |
ENST00000498416.2
ENST00000465066.5 |
DIAPH3
|
diaphanous related formin 3 |
| chr18_+_74534594 | 0.22 |
ENST00000582365.1
|
CNDP1
|
carnosine dipeptidase 1 |
| chr18_-_3845292 | 0.20 |
ENST00000400145.6
|
DLGAP1
|
DLG associated protein 1 |
| chr19_+_48695952 | 0.20 |
ENST00000522966.2
ENST00000425340.3 ENST00000391876.5 |
FUT2
|
fucosyltransferase 2 |
| chr1_+_115029823 | 0.19 |
ENST00000256592.3
|
TSHB
|
thyroid stimulating hormone subunit beta |
| chr1_+_197268204 | 0.19 |
ENST00000535699.5
ENST00000538660.5 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr4_+_157220691 | 0.17 |
ENST00000509417.5
ENST00000645636.1 ENST00000296526.12 ENST00000264426.14 |
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr5_-_16916400 | 0.14 |
ENST00000513882.5
|
MYO10
|
myosin X |
| chr13_-_49401497 | 0.08 |
ENST00000457041.5
ENST00000355854.8 |
CAB39L
|
calcium binding protein 39 like |
| chrX_+_85003863 | 0.08 |
ENST00000373173.7
|
APOOL
|
apolipoprotein O like |
| chr4_-_23890035 | 0.07 |
ENST00000507380.1
ENST00000264867.7 |
PPARGC1A
|
PPARG coactivator 1 alpha |
| chr19_+_48445961 | 0.05 |
ENST00000253237.10
|
GRWD1
|
glutamate rich WD repeat containing 1 |
| chr12_-_86256299 | 0.04 |
ENST00000552808.6
ENST00000547225.5 |
MGAT4C
|
MGAT4 family member C |
| chr7_+_120273129 | 0.02 |
ENST00000331113.9
|
KCND2
|
potassium voltage-gated channel subfamily D member 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 1.1 | 3.3 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.6 | 1.9 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.6 | 3.0 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.5 | 2.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.5 | 1.8 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.4 | 2.4 | GO:0070101 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.4 | 5.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.4 | 2.9 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.3 | 2.4 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.3 | 17.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.3 | 6.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.3 | 1.6 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.2 | 1.5 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 4.6 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.2 | 2.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 1.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 2.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 0.5 | GO:0036304 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 0.2 | 3.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.7 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.1 | 3.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.5 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 3.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.8 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.9 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 1.2 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 0.9 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 1.4 | GO:1900103 | NFAT protein import into nucleus(GO:0051531) positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.1 | 2.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.5 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 4.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.4 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.1 | 2.9 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 2.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 4.4 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 1.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 1.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.2 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 1.1 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 3.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 17.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.7 | 6.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.3 | 1.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.3 | 1.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.2 | 1.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 4.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.1 | 0.7 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 2.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.2 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 2.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 3.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 2.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 4.3 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 1.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.6 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 2.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 3.0 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.1 | GO:0035579 | specific granule membrane(GO:0035579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 1.7 | 5.0 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 1.0 | 6.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 1.0 | 3.0 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.8 | 3.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.7 | 3.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.5 | 3.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.3 | 2.4 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.3 | 17.3 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.3 | 2.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 1.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.2 | 0.5 | GO:0042806 | fucose binding(GO:0042806) |
| 0.2 | 4.6 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.2 | 1.4 | GO:0046935 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.5 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 2.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 1.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 4.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 3.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 3.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 1.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.3 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 1.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 2.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.4 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 1.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 2.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 3.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 3.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.7 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 3.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 4.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 5.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 3.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 3.7 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 1.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 4.4 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 1.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 3.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 6.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.9 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 1.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |