Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for LMX1B_MNX1_RAX2
Z-value: 2.75
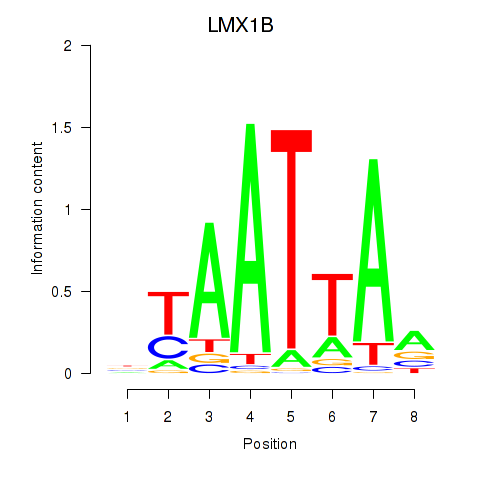
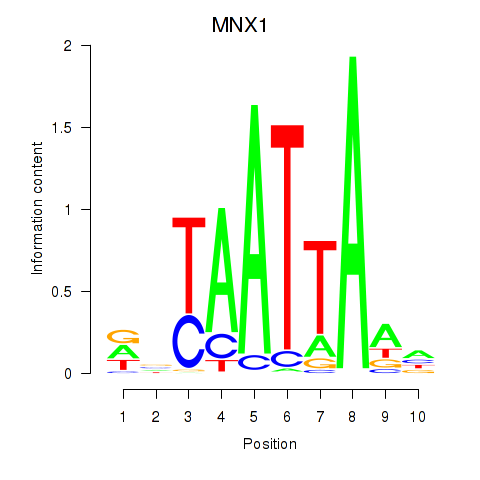
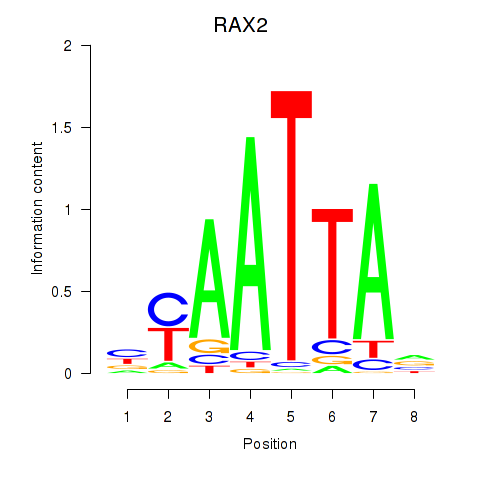
Transcription factors associated with LMX1B_MNX1_RAX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LMX1B
|
ENSG00000136944.19 | LMX1B |
|
MNX1
|
ENSG00000130675.15 | MNX1 |
|
RAX2
|
ENSG00000173976.16 | RAX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LMX1B | hg38_v1_chr9_+_126613922_126613946 | -0.15 | 2.7e-02 | Click! |
| RAX2 | hg38_v1_chr19_-_3772211_3772238 | -0.08 | 2.3e-01 | Click! |
| MNX1 | hg38_v1_chr7_-_157010615_157010668 | 0.07 | 2.9e-01 | Click! |
Activity profile of LMX1B_MNX1_RAX2 motif
Sorted Z-values of LMX1B_MNX1_RAX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LMX1B_MNX1_RAX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 3.3 | 62.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 1.8 | 7.2 | GO:0090427 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 1.6 | 4.8 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 1.5 | 7.6 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 1.1 | 6.5 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 1.0 | 5.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.9 | 2.7 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.7 | 9.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.7 | 2.9 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.6 | 2.5 | GO:0098923 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.6 | 1.8 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.6 | 7.9 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.5 | 6.5 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.5 | 2.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.5 | 5.6 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.5 | 1.8 | GO:2000426 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.5 | 4.1 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.5 | 1.8 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.4 | 4.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.4 | 2.9 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.4 | 2.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.3 | 2.4 | GO:0090269 | fibroblast growth factor production(GO:0090269) regulation of fibroblast growth factor production(GO:0090270) |
| 0.3 | 8.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 1.6 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.3 | 3.4 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.3 | 3.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.3 | 4.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.3 | 9.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.3 | 2.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.2 | 0.7 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.2 | 2.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 0.6 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.2 | 3.9 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 2.0 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 0.6 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.2 | 0.8 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.2 | 0.6 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.2 | 9.9 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.2 | 3.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.2 | 1.9 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.2 | 1.5 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.2 | 0.6 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.2 | 7.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 1.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 1.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.1 | 6.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.5 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 0.5 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.1 | 0.5 | GO:2000354 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.6 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.1 | 8.7 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 5.4 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 0.6 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 1.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.8 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 0.6 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.4 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 2.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 1.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.7 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 2.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 2.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.8 | GO:0060353 | regulation of cell adhesion molecule production(GO:0060353) positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 1.0 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 2.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 3.5 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 1.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.7 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 1.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 1.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 3.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 0.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 1.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 1.1 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 0.4 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.7 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.8 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 1.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:1903961 | positive regulation of anion channel activity(GO:1901529) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 2.7 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 4.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 3.1 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.6 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.2 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.4 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 2.3 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 1.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 2.9 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 | 0.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 1.2 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) |
| 0.0 | 0.6 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 1.0 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.9 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.3 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 2.3 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 3.0 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 3.6 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 1.1 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.3 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.2 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 2.5 | 7.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.5 | 22.4 | GO:0030478 | actin cap(GO:0030478) |
| 1.2 | 62.1 | GO:0002102 | podosome(GO:0002102) |
| 1.2 | 6.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.8 | 3.9 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.6 | 1.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.4 | 2.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.3 | 1.8 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.3 | 12.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 4.7 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 2.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 3.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 2.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 3.6 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.2 | 0.7 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.2 | 12.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 2.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 2.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.7 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.5 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.7 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 2.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 4.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 3.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.3 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 1.6 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 3.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 5.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 9.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 10.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 3.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 14.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 15.7 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 6.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.0 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.0 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 2.7 | 63.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 2.0 | 9.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.7 | 5.0 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 1.0 | 4.8 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.8 | 22.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.7 | 3.6 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.6 | 2.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.6 | 2.8 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.5 | 3.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.5 | 2.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.5 | 3.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.4 | 8.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.4 | 1.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.4 | 2.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.4 | 2.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 1.0 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.3 | 1.8 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 1.8 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 2.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 3.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 2.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 6.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 2.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 3.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 3.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.2 | 3.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 0.5 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.2 | 1.0 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.2 | 0.8 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 0.8 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.6 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 2.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.7 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 0.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 2.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 3.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.7 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.8 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 1.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 2.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 1.9 | GO:0030297 | ErbB-2 class receptor binding(GO:0005176) transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 1.0 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 9.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 1.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 8.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.5 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 4.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 3.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 5.0 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 3.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 4.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 2.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 7.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 11.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 1.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 1.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.4 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 3.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 4.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 5.4 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 11.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 7.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 9.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 8.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 0.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 4.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 5.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 7.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 7.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.3 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.5 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.5 | 8.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.4 | 24.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 8.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 3.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 3.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 2.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 5.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 3.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 6.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 7.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 3.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 2.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 4.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 3.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 4.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 3.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 4.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 3.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |