Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
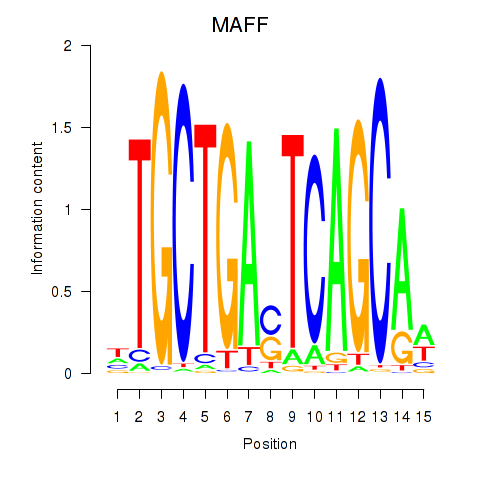
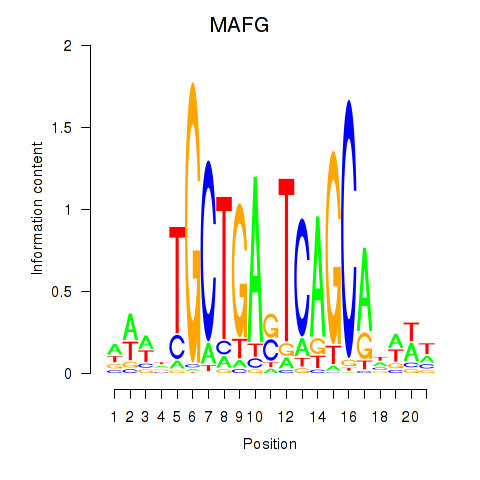
Results for MAFF_MAFG
Z-value: 4.67
Transcription factors associated with MAFF_MAFG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFF
|
ENSG00000185022.12 | MAFF |
|
MAFG
|
ENSG00000197063.11 | MAFG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFG | hg38_v1_chr17_-_81923532_81923547, hg38_v1_chr17_-_81927699_81927748 | 0.10 | 1.2e-01 | Click! |
| MAFF | hg38_v1_chr22_+_38201932_38202014 | 0.04 | 5.3e-01 | Click! |
Activity profile of MAFF_MAFG motif
Sorted Z-values of MAFF_MAFG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFF_MAFG
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 2.7 | 8.0 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 2.1 | 8.6 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 1.7 | 13.3 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 1.6 | 4.8 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) protection from natural killer cell mediated cytotoxicity(GO:0042270) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 1.3 | 3.8 | GO:0072275 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 1.2 | 6.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 1.2 | 9.6 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.0 | 7.8 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 1.0 | 9.5 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.9 | 3.6 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.9 | 2.6 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.8 | 4.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.7 | 4.0 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.6 | 6.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.6 | 1.9 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.6 | 1.8 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.6 | 1.7 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.6 | 1.7 | GO:1901097 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of autophagosome maturation(GO:1901097) |
| 0.6 | 1.7 | GO:0018307 | tRNA wobble position uridine thiolation(GO:0002143) enzyme active site formation(GO:0018307) |
| 0.5 | 1.6 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.5 | 2.1 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.5 | 3.0 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.5 | 4.9 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.5 | 2.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.5 | 1.4 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.5 | 1.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.5 | 2.3 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.4 | 67.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.4 | 1.2 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.4 | 2.0 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.4 | 56.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.4 | 3.5 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.4 | 8.8 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.4 | 2.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.4 | 3.8 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.4 | 2.2 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.4 | 1.8 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 | 1.0 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.3 | 1.4 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.3 | 1.0 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.3 | 3.6 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 5.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 1.9 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.3 | 1.3 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.3 | 5.6 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.3 | 21.1 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.3 | 3.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.3 | 2.5 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.3 | 0.6 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.3 | 4.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 5.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.3 | 2.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.3 | 1.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 2.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.2 | 2.9 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 2.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 1.8 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.2 | 0.9 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.2 | 1.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 0.8 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 2.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 8.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 1.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 1.6 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 1.0 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.2 | 1.3 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.2 | 1.4 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.2 | 4.7 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.2 | 2.9 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.2 | 1.2 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.2 | 1.5 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 0.5 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.2 | 1.0 | GO:0010044 | response to aluminum ion(GO:0010044) |
| 0.1 | 0.9 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 0.6 | GO:0052148 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.1 | 5.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 4.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 4.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 9.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 7.8 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.1 | 0.5 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 0.5 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.3 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 20.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.4 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 1.4 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.4 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 1.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 2.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.7 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 2.0 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 1.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.8 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.4 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 7.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 1.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.4 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.3 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 2.0 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.6 | GO:0035356 | positive regulation of sequestering of triglyceride(GO:0010890) cellular triglyceride homeostasis(GO:0035356) |
| 0.1 | 1.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.2 | GO:0045359 | dendritic cell proliferation(GO:0044565) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 1.5 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 1.2 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.1 | 3.6 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 0.5 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 3.1 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.1 | 0.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.5 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 2.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 4.4 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.6 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.8 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 0.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 5.8 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 1.3 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.1 | 0.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 2.3 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 17.9 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 2.0 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 1.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.5 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.7 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 2.7 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 0.9 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 1.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 13.8 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 0.4 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 1.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 2.8 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.1 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.9 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 1.6 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.4 | GO:0002775 | antimicrobial peptide production(GO:0002775) antibacterial peptide production(GO:0002778) |
| 0.0 | 11.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.8 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 2.1 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 1.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 2.7 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 | 3.6 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 2.2 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.4 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 1.4 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 1.9 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.3 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 2.0 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 1.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.7 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 2.2 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.5 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.1 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.2 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 1.0 | GO:0008033 | tRNA processing(GO:0008033) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 20.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 3.7 | 15.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 3.0 | 8.9 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 2.1 | 8.6 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 1.2 | 77.7 | GO:0005844 | polysome(GO:0005844) |
| 1.1 | 21.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.8 | 6.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.6 | 11.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.5 | 3.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.5 | 3.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.5 | 1.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.5 | 5.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 4.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.4 | 2.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 3.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.4 | 2.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.4 | 7.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 1.5 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.3 | 3.8 | GO:0042611 | MHC protein complex(GO:0042611) MHC class I protein complex(GO:0042612) |
| 0.3 | 3.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 3.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.3 | 2.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.3 | 4.1 | GO:0033177 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 1.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 0.9 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 0.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 9.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 0.8 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.2 | 2.0 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 3.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.6 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 5.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 2.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 4.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 4.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 2.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 3.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 7.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 2.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 5.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.9 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 15.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 11.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 7.8 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 5.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 8.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 2.9 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 9.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 4.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 6.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 3.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.3 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 6.0 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 2.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 3.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 2.2 | GO:0070603 | SWI/SNF superfamily-type complex(GO:0070603) |
| 0.0 | 1.1 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 1.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 6.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.2 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.9 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 22.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.1 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 3.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 15.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 1.9 | 9.5 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 1.6 | 6.5 | GO:0047291 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.5 | 13.3 | GO:0043426 | MRF binding(GO:0043426) |
| 1.3 | 3.8 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.3 | 6.4 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 1.1 | 5.6 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 1.1 | 8.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.1 | 85.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 1.0 | 20.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.8 | 4.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.8 | 0.8 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.8 | 4.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.8 | 9.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.7 | 27.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.7 | 1.4 | GO:0050473 | arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.5 | 3.3 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.5 | 3.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.5 | 8.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.5 | 2.0 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.5 | 5.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.5 | 1.9 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.5 | 7.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.5 | 5.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.5 | 1.4 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 1.7 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.4 | 1.7 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.4 | 4.0 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.4 | 5.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.4 | 1.2 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.3 | 1.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 1.0 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.3 | 1.0 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.3 | 65.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.3 | 1.3 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.3 | 4.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.3 | 8.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.3 | 4.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 2.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.3 | 4.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 8.5 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.3 | 4.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 1.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.3 | 2.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 2.1 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.3 | 7.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 3.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 1.0 | GO:0070404 | NADH binding(GO:0070404) |
| 0.2 | 1.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 0.7 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 4.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 2.7 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.2 | 2.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 4.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.2 | 0.6 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.2 | 2.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 6.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 0.5 | GO:0052852 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.2 | 0.5 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.2 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.9 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 0.7 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 2.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.9 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 1.4 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.1 | 1.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 4.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.0 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 8.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 1.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.7 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.1 | 0.4 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 2.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 1.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.3 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.1 | 0.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.7 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 21.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 5.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 2.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 4.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.4 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.4 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 2.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 2.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.9 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 1.0 | GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism(GO:0015662) |
| 0.1 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 3.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 1.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 6.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.3 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 1.0 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 1.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 1.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.2 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.6 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 1.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.6 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 2.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0022821 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 1.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 1.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 15.6 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 14.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 11.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 15.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 12.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 30.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 20.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 3.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 4.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 4.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 4.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.8 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 20.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.0 | 21.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 1.0 | 76.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.4 | 5.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 8.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.3 | 10.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.3 | 21.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 6.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 4.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 5.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 3.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 2.9 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.2 | 2.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 4.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 2.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 16.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 4.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 2.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.8 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 3.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 5.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 1.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 21.3 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.9 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 1.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |