Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
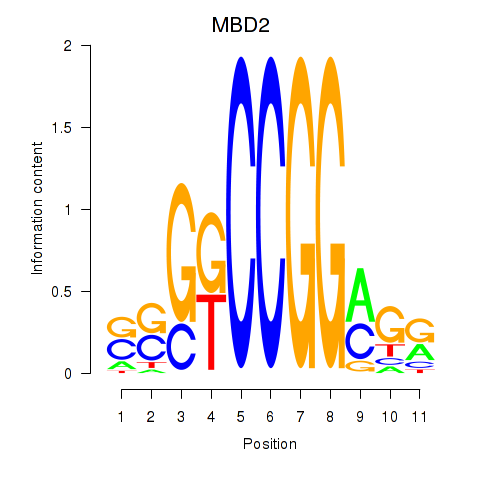
Results for MBD2
Z-value: 0.28
Transcription factors associated with MBD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MBD2
|
ENSG00000134046.12 | MBD2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MBD2 | hg38_v1_chr18_-_54224578_54224689 | -0.10 | 1.5e-01 | Click! |
Activity profile of MBD2 motif
Sorted Z-values of MBD2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MBD2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.8 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 1.5 | 4.6 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 1.5 | 6.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 1.3 | 9.4 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 1.3 | 4.0 | GO:0060596 | mammary placode formation(GO:0060596) His-Purkinje system cell differentiation(GO:0060932) |
| 1.2 | 6.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 1.2 | 8.6 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 1.2 | 3.6 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 1.2 | 5.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 1.1 | 6.8 | GO:0035602 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 1.0 | 4.0 | GO:0061643 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.9 | 4.7 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.9 | 3.6 | GO:0019086 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.8 | 1.6 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.8 | 2.4 | GO:0071072 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) negative regulation of phospholipid biosynthetic process(GO:0071072) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.8 | 6.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.8 | 3.9 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.8 | 2.3 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.7 | 2.2 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.7 | 5.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.7 | 2.2 | GO:0060528 | regulation of translational initiation by iron(GO:0006447) secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.7 | 2.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.7 | 4.8 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.7 | 5.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.7 | 1.3 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.7 | 2.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.6 | 3.8 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.6 | 6.0 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.6 | 6.3 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.6 | 1.1 | GO:0030806 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) |
| 0.5 | 6.5 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.5 | 2.6 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.5 | 2.6 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.5 | 1.5 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.5 | 4.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.5 | 2.1 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.5 | 2.5 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.5 | 1.5 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.5 | 3.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.4 | 2.6 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.4 | 2.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.4 | 1.3 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.4 | 2.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.4 | 1.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.4 | 1.5 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.4 | 5.0 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.4 | 3.6 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.4 | 2.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 1.7 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.3 | 4.8 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.3 | 3.0 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 1.0 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.3 | 3.3 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.3 | 2.3 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.3 | 2.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 2.1 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.3 | 1.8 | GO:0051532 | regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.3 | 1.5 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.3 | 2.9 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.3 | 1.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.3 | 2.3 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.3 | 1.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.3 | 1.1 | GO:1904021 | regulation of G-protein coupled receptor internalization(GO:1904020) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.3 | 1.4 | GO:0090269 | fibroblast growth factor production(GO:0090269) regulation of fibroblast growth factor production(GO:0090270) |
| 0.3 | 5.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.3 | 3.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 0.8 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.3 | 1.0 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.3 | 1.3 | GO:0072093 | metanephric renal vesicle formation(GO:0072093) |
| 0.3 | 1.3 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.3 | 1.8 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 1.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 | 2.9 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.2 | 2.7 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.2 | 6.4 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.2 | 2.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 2.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.2 | 0.9 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.2 | 2.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 6.7 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.2 | 4.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.2 | 4.5 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.2 | 1.0 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 1.0 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 0.2 | 3.1 | GO:0060736 | positive regulation of glial cell proliferation(GO:0060252) prostate gland growth(GO:0060736) |
| 0.2 | 0.8 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.2 | 0.4 | GO:0060437 | lung growth(GO:0060437) |
| 0.2 | 2.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 8.7 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.2 | 2.0 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 2.0 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.2 | 0.9 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.2 | 2.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 0.5 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 7.1 | GO:0098534 | centriole assembly(GO:0098534) |
| 0.2 | 1.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 0.9 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 0.8 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 0.3 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.2 | 2.9 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.2 | 1.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 2.4 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 2.2 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.2 | 3.7 | GO:0090494 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.2 | 3.6 | GO:0098915 | potassium ion export across plasma membrane(GO:0097623) membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.2 | 0.9 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.2 | 2.9 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.2 | 2.9 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 0.3 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.1 | 1.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.5 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 2.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 3.2 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 1.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 1.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.4 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.9 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 1.9 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 1.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 3.8 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 3.0 | GO:0036120 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 2.0 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.1 | 2.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.4 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 2.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.6 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 1.7 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.1 | 1.8 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 0.3 | GO:0031550 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.1 | 2.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 2.2 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.1 | 1.5 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.1 | 1.6 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 1.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.1 | 1.4 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.1 | 3.3 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) |
| 0.1 | 0.9 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.6 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 1.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 11.2 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.1 | 2.0 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.9 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 0.9 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.1 | 6.0 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 0.8 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 4.2 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 3.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.1 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 2.0 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.1 | 0.1 | GO:0032687 | regulation of interleukin-3 production(GO:0032672) negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 0.8 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 6.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 1.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 0.2 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 1.6 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 3.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 2.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.1 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.7 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.9 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.1 | 1.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.8 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 1.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.1 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 0.2 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) positive regulation of microtubule binding(GO:1904528) |
| 0.1 | 1.4 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 0.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.0 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.1 | 2.5 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 2.0 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.9 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 2.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 2.1 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 1.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 12.1 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 3.0 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 2.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0070525 | tRNA threonylcarbamoyladenosine modification(GO:0002949) tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 1.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 1.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 3.6 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.0 | 1.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 1.7 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.6 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 2.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.8 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 1.8 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 1.0 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.9 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 2.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 2.8 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 1.4 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 3.1 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.5 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.1 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) Schwann cell proliferation(GO:0014010) |
| 0.0 | 1.0 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 1.4 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 1.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 3.1 | GO:0001505 | regulation of neurotransmitter levels(GO:0001505) |
| 0.0 | 0.7 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 2.2 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.5 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 1.3 | GO:0044782 | cilium organization(GO:0044782) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.8 | 4.9 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.7 | 8.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.7 | 2.2 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.7 | 4.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.6 | 3.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.5 | 5.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.4 | 1.8 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.4 | 1.9 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.3 | 9.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.3 | 3.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.3 | 1.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 5.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 1.5 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 5.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 3.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 1.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.2 | 4.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 4.0 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 6.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.2 | 14.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 2.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 1.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 6.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 0.9 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 1.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.5 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 2.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 1.6 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 1.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 3.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 3.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.9 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 1.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 6.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 0.6 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 0.6 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 1.2 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.1 | 1.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 4.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 0.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.7 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 1.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 6.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 1.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 8.0 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 3.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 7.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 2.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 3.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 3.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.1 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 2.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 5.4 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 1.1 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 1.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 3.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 2.0 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 43.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.3 | GO:0070160 | occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 8.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.9 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.2 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.1 | GO:0044450 | microtubule organizing center part(GO:0044450) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0046577 | long-chain-alcohol oxidase activity(GO:0046577) |
| 1.7 | 5.0 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 1.5 | 5.8 | GO:0001626 | nociceptin receptor activity(GO:0001626) |
| 1.4 | 5.5 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 1.3 | 9.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 1.3 | 5.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.1 | 3.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 1.1 | 4.3 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 1.0 | 5.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.9 | 3.7 | GO:0005334 | norepinephrine:sodium symporter activity(GO:0005334) |
| 0.8 | 9.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.8 | 2.4 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.8 | 4.0 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.7 | 8.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.7 | 2.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.7 | 5.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.7 | 2.0 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.6 | 3.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.6 | 4.9 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.6 | 7.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.6 | 4.0 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.6 | 4.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.6 | 2.2 | GO:0070644 | 9-cis retinoic acid receptor activity(GO:0004886) vitamin D response element binding(GO:0070644) |
| 0.5 | 3.8 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.5 | 1.6 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.5 | 3.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.5 | 6.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.5 | 3.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.5 | 2.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.5 | 4.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.4 | 2.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 3.0 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.4 | 5.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 5.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.4 | 3.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.4 | 1.1 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.4 | 1.5 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.4 | 2.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 1.4 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.3 | 9.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.3 | 3.7 | GO:0036122 | BMP binding(GO:0036122) |
| 0.3 | 3.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 1.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.3 | 0.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.3 | 2.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 2.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 2.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 1.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 12.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.3 | 1.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.3 | 0.8 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.3 | 1.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 2.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 4.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.3 | 3.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.3 | 1.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 2.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.2 | 2.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.6 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.2 | 1.6 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.2 | 0.7 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 1.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 0.6 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.2 | 3.0 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 0.9 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 5.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 0.5 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 2.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 2.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 5.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 1.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.2 | 1.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 1.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 4.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 1.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 3.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 7.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 1.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 5.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 16.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.6 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) L-phenylalanine aminotransferase activity(GO:0070546) |
| 0.1 | 1.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 2.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.8 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 2.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.9 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 1.3 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 1.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 2.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 1.9 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 5.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 2.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.9 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 4.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 5.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 1.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 1.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 1.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 2.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 4.2 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 2.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 2.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 1.0 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 5.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 3.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 3.3 | GO:0008235 | aminopeptidase activity(GO:0004177) metalloexopeptidase activity(GO:0008235) |
| 0.1 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.6 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 1.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 1.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 2.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 11.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 2.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 1.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 2.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 3.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 1.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 18.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 9.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.3 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 1.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 6.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.9 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 4.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 2.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 2.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 7.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 2.2 | GO:0008168 | methyltransferase activity(GO:0008168) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 8.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 6.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 6.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 3.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 2.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 9.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 5.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 7.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 10.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 0.9 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 8.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 11.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 7.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 5.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 5.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 4.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 4.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 4.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.3 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 3.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 4.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 3.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 2.9 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.5 | 8.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 6.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.4 | 2.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.4 | 9.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 1.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.3 | 2.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 6.6 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.3 | 7.7 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.2 | 17.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 7.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 7.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 5.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 2.8 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 2.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 3.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 1.1 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.1 | 2.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 11.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 6.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 10.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 4.8 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 2.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 4.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 0.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 3.9 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 2.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 2.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.7 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 2.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 4.5 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 9.2 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 1.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 2.1 | REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | Genes involved in Signaling by TGF-beta Receptor Complex |
| 0.0 | 2.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 2.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.0 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.8 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |