Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
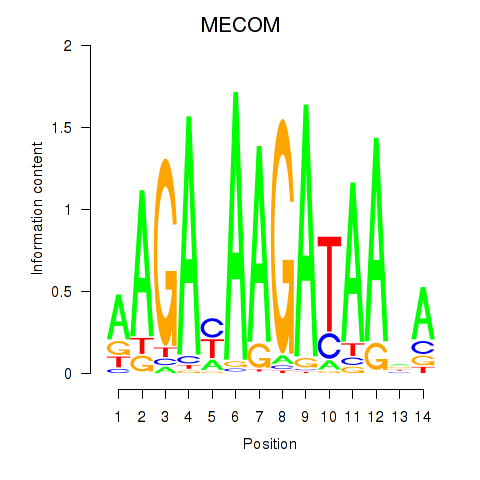
Results for MECOM
Z-value: 7.39
Transcription factors associated with MECOM
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MECOM
|
ENSG00000085276.19 | MECOM |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MECOM | hg38_v1_chr3_-_169146595_169146675, hg38_v1_chr3_-_169147734_169147748, hg38_v1_chr3_-_169663704_169663775 | -0.03 | 6.9e-01 | Click! |
Activity profile of MECOM motif
Sorted Z-values of MECOM motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MECOM
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.5 | 34.6 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 4.3 | 12.8 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 4.2 | 8.4 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 3.9 | 19.5 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 3.5 | 10.6 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 3.5 | 10.5 | GO:0018963 | phthalate metabolic process(GO:0018963) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 3.1 | 9.2 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 2.5 | 10.1 | GO:1903296 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 2.4 | 30.9 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 1.9 | 17.2 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 1.9 | 7.6 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 1.6 | 9.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.6 | 9.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 1.4 | 9.8 | GO:0010044 | response to aluminum ion(GO:0010044) |
| 1.4 | 5.5 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 1.3 | 5.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 1.3 | 3.8 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 1.2 | 11.8 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 1.1 | 3.4 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 1.0 | 3.0 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.0 | 7.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.9 | 3.7 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.9 | 5.5 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.9 | 18.7 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.9 | 3.6 | GO:2001034 | histone H3-K36 dimethylation(GO:0097676) positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.9 | 25.5 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.9 | 3.4 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.8 | 8.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.7 | 5.9 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.7 | 2.9 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.7 | 2.1 | GO:0035425 | autocrine signaling(GO:0035425) canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.7 | 4.7 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.6 | 5.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.6 | 6.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.6 | 5.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.6 | 7.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.6 | 7.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.5 | 12.4 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.5 | 1.4 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.5 | 2.3 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.4 | 4.0 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.4 | 2.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.4 | 8.5 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.4 | 6.9 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.4 | 1.9 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.4 | 9.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.4 | 16.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.3 | 1.7 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 1.7 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.3 | 1.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 2.3 | GO:0060011 | progesterone biosynthetic process(GO:0006701) Sertoli cell proliferation(GO:0060011) |
| 0.3 | 2.9 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.3 | 2.3 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 1.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 2.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.3 | 4.5 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.3 | 6.0 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.3 | 9.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.3 | 5.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.3 | 6.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.3 | 26.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 16.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.2 | 4.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 3.0 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 1.0 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 6.9 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.2 | 2.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.2 | 3.7 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.2 | 9.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 0.9 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 6.8 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 0.8 | GO:0072501 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 2.3 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.1 | 12.9 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 5.0 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 1.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 10.5 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.1 | 4.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 8.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 2.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 2.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 2.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 3.9 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 2.9 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 1.9 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 2.8 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 5.2 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 4.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 2.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 3.7 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 4.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.9 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 20.4 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 7.4 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.9 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 1.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 2.1 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 4.1 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 2.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 2.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 3.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 3.9 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 10.1 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 3.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.3 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 1.1 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 1.5 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 1.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.7 | 34.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.7 | 10.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 1.3 | 5.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 1.3 | 5.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 1.2 | 3.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.0 | 10.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.9 | 5.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.8 | 3.0 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.8 | 9.8 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.7 | 10.3 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.7 | 2.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.7 | 3.4 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.6 | 3.5 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.5 | 3.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.5 | 7.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.5 | 5.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 4.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.4 | 7.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 3.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.3 | 4.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 10.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.3 | 5.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 2.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 93.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.2 | 9.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 2.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 2.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 3.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 9.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 3.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 3.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 7.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 21.2 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 2.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 2.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 6.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 7.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 12.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 1.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 9.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 4.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 4.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 4.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 8.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 4.8 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 3.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 110.5 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.9 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 2.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 17.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 34.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 4.3 | 12.9 | GO:0030305 | heparanase activity(GO:0030305) |
| 3.4 | 17.2 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 2.8 | 8.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 2.8 | 30.9 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 1.8 | 9.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.7 | 5.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 1.4 | 6.9 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 1.4 | 5.5 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 1.3 | 12.8 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 1.2 | 3.7 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.2 | 3.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.2 | 19.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 1.1 | 5.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 1.0 | 9.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.9 | 7.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.9 | 6.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.8 | 7.9 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.8 | 2.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.8 | 9.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.7 | 5.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.7 | 5.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.7 | 4.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.6 | 13.0 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.5 | 2.9 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.5 | 7.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 3.6 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.4 | 1.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.4 | 3.0 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.4 | 6.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.4 | 35.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 4.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.4 | 3.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.4 | 10.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.4 | 2.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.4 | 2.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.7 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.3 | 9.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 0.9 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.3 | 6.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 12.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 9.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 26.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.3 | 3.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.3 | 7.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.3 | 4.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.3 | 4.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 2.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 4.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 18.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 8.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 27.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 1.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 7.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 2.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 3.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 2.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 19.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 1.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 2.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.0 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 2.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 8.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 3.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 2.6 | GO:0042166 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.1 | 1.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 5.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 4.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 8.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 12.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 1.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 2.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.0 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 15.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 4.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.6 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 3.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.1 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 2.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 9.2 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 16.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 71.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 6.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 10.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 16.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 6.9 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 2.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 19.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 6.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 25.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 6.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 12.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 4.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 4.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 6.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 4.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 4.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 11.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.9 | 29.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.8 | 34.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.8 | 14.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.7 | 8.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.5 | 3.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.5 | 12.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.5 | 10.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.4 | 14.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.4 | 13.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 3.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 9.7 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 18.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 17.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 3.8 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 3.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 29.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 3.9 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 9.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 3.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 11.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 10.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 4.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 7.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 14.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 3.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 4.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 4.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 2.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.9 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |