Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for MEF2D_MEF2A
Z-value: 8.31
Transcription factors associated with MEF2D_MEF2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
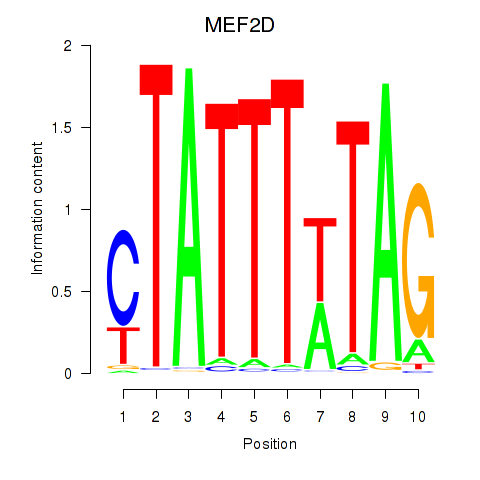
MEF2D
|
ENSG00000116604.18 | MEF2D |
|
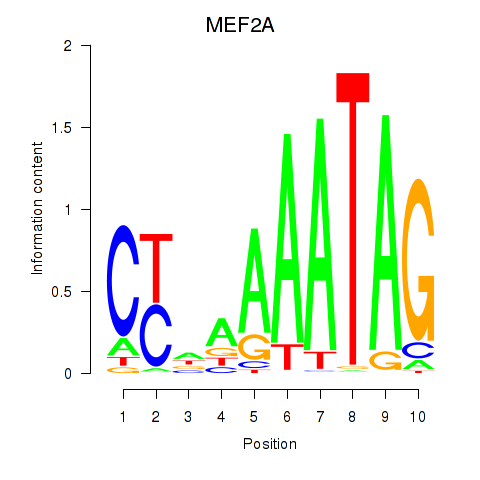
MEF2A
|
ENSG00000068305.17 | MEF2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2D | hg38_v1_chr1_-_156490599_156490625, hg38_v1_chr1_-_156500763_156500851 | 0.54 | 3.2e-18 | Click! |
| MEF2A | hg38_v1_chr15_+_99565950_99566038 | 0.23 | 7.4e-04 | Click! |
Activity profile of MEF2D_MEF2A motif
Sorted Z-values of MEF2D_MEF2A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2D_MEF2A
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.4 | 92.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 14.3 | 71.3 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 9.4 | 28.3 | GO:1900920 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 9.2 | 27.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 7.9 | 47.3 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 7.5 | 22.4 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 6.8 | 116.1 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 5.9 | 17.7 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 5.0 | 60.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 4.8 | 52.7 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 4.7 | 14.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 4.4 | 21.8 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 4.3 | 60.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 3.8 | 15.3 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 3.4 | 31.0 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 3.3 | 16.3 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 3.2 | 12.6 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 3.1 | 62.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 3.0 | 11.8 | GO:0032972 | regulation of muscle filament sliding(GO:0032971) regulation of muscle filament sliding speed(GO:0032972) |
| 2.9 | 17.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 2.9 | 26.0 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 2.9 | 23.0 | GO:0061709 | reticulophagy(GO:0061709) |
| 2.7 | 18.9 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 2.5 | 7.5 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 2.4 | 19.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 2.4 | 16.8 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 2.3 | 9.2 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 2.1 | 19.0 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 2.1 | 46.2 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 2.0 | 9.9 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 2.0 | 117.1 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 2.0 | 9.8 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.9 | 11.7 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 1.9 | 5.8 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 1.9 | 32.8 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 1.9 | 24.8 | GO:1902548 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 1.9 | 5.7 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 1.9 | 11.3 | GO:0015853 | adenine transport(GO:0015853) |
| 1.8 | 17.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.7 | 29.7 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 1.6 | 6.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 1.5 | 5.9 | GO:0036369 | traversing start control point of mitotic cell cycle(GO:0007089) transcription factor catabolic process(GO:0036369) cellular response to actinomycin D(GO:0072717) |
| 1.4 | 4.3 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 1.4 | 5.6 | GO:0015793 | glycerol transport(GO:0015793) |
| 1.3 | 3.9 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 1.3 | 7.7 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 1.3 | 6.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.2 | 12.3 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 1.2 | 13.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 1.2 | 3.5 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 1.2 | 23.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.2 | 3.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 1.1 | 9.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 1.0 | 4.9 | GO:2000670 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.9 | 42.6 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.9 | 2.8 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.9 | 15.8 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.9 | 2.8 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.9 | 16.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.9 | 36.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.8 | 6.8 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.8 | 12.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.8 | 12.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.8 | 20.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.8 | 2.3 | GO:0070256 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.7 | 21.2 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.7 | 5.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.7 | 12.0 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.7 | 2.8 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.7 | 11.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.7 | 6.7 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.7 | 51.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.6 | 4.8 | GO:0007512 | adult heart development(GO:0007512) |
| 0.6 | 7.6 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.5 | 5.3 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.5 | 4.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.5 | 3.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.5 | 4.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.5 | 12.7 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.5 | 70.8 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.5 | 4.3 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.5 | 12.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.5 | 6.0 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.5 | 3.6 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.5 | 1.8 | GO:1902938 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.4 | 5.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.4 | 16.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.4 | 10.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.4 | 2.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.4 | 2.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.4 | 1.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.4 | 1.5 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.4 | 3.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.4 | 12.7 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.4 | 12.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.4 | 2.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.3 | 12.6 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.3 | 2.4 | GO:1990822 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.3 | 63.2 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.3 | 0.7 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.3 | 5.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 27.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.3 | 2.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.3 | 5.3 | GO:0010611 | regulation of cardiac muscle hypertrophy(GO:0010611) |
| 0.3 | 4.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.3 | 8.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 4.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 0.9 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.3 | 14.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.3 | 0.8 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.3 | 19.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.3 | 6.1 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 | 18.1 | GO:0061045 | negative regulation of wound healing(GO:0061045) |
| 0.2 | 28.7 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.2 | 9.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.2 | 20.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 33.5 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.2 | 3.5 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 1.6 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.2 | 1.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 4.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.2 | 2.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 8.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 13.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 1.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 0.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 9.3 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.2 | 6.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 3.0 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.2 | 4.9 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.2 | 0.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 3.5 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.2 | 8.1 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.2 | 12.0 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.2 | 11.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.2 | 10.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 5.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.9 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 5.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.4 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.1 | 1.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 6.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 4.0 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 4.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 3.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.8 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 12.7 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.1 | 4.3 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 1.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 1.6 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 4.7 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 2.0 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 2.0 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 2.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.1 | 8.0 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 1.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.2 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 0.8 | GO:1903392 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 5.7 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.1 | 0.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 13.1 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.1 | 11.9 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 4.7 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 1.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 7.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.4 | GO:0048265 | response to pain(GO:0048265) |
| 0.0 | 0.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 2.3 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.0 | 3.6 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 7.2 | GO:0046578 | regulation of Ras protein signal transduction(GO:0046578) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.8 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) |
| 0.0 | 5.0 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 2.1 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 37.8 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 5.5 | 27.5 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 5.3 | 21.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 4.9 | 78.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 4.2 | 75.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 3.8 | 15.3 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 3.7 | 63.2 | GO:0005861 | troponin complex(GO:0005861) |
| 3.1 | 12.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 2.6 | 103.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 2.3 | 15.8 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 1.9 | 24.8 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.8 | 27.6 | GO:0030478 | actin cap(GO:0030478) |
| 1.8 | 23.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.7 | 24.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 1.5 | 12.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.3 | 12.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.3 | 16.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 1.2 | 13.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.1 | 5.7 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.0 | 59.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 1.0 | 19.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.9 | 15.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.9 | 9.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.8 | 176.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.8 | 17.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.8 | 157.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.8 | 49.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.7 | 2.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.5 | 51.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.5 | 2.7 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.5 | 29.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.4 | 15.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.4 | 23.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.4 | 14.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.3 | 12.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 28.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 57.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.3 | 5.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 3.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 9.8 | GO:0071564 | SWI/SNF complex(GO:0016514) npBAF complex(GO:0071564) |
| 0.3 | 2.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 28.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 0.7 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.2 | 51.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 6.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 11.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 43.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 10.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 1.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 8.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 2.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 6.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 19.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 5.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 1.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 3.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 7.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 4.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 32.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 3.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 9.8 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 8.1 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 6.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 7.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 23.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 3.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 8.4 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.1 | 2.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 3.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 4.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 18.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.3 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 26.7 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 4.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 8.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.8 | GO:0032587 | ruffle membrane(GO:0032587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.5 | 62.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 10.3 | 20.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 10.2 | 30.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 5.9 | 17.7 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 5.5 | 27.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 5.4 | 43.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 5.3 | 58.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 5.2 | 21.0 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 4.5 | 26.7 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 4.2 | 62.3 | GO:0031432 | titin binding(GO:0031432) |
| 3.8 | 15.3 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 3.8 | 75.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 3.7 | 52.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 3.5 | 14.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 3.5 | 49.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 3.3 | 29.7 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 3.2 | 12.6 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 2.9 | 11.7 | GO:0030172 | troponin C binding(GO:0030172) |
| 2.8 | 11.3 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 2.7 | 11.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 2.7 | 21.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 2.6 | 48.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 2.6 | 7.7 | GO:0070538 | oleic acid binding(GO:0070538) |
| 2.5 | 7.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 2.5 | 9.9 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 2.3 | 11.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 2.3 | 11.7 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 2.2 | 28.3 | GO:1990239 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 2.1 | 31.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.9 | 73.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 1.7 | 11.9 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 1.7 | 8.4 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 1.6 | 19.4 | GO:0008430 | selenium binding(GO:0008430) |
| 1.6 | 6.4 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 1.6 | 98.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 1.5 | 12.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.5 | 21.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 1.4 | 12.3 | GO:0046935 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 1.3 | 6.5 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 1.3 | 3.9 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 1.2 | 3.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 1.2 | 10.8 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 1.2 | 8.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.0 | 17.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.0 | 3.0 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.9 | 12.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.9 | 12.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.8 | 5.9 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.8 | 3.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.8 | 23.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.7 | 6.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.7 | 3.6 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.7 | 16.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.7 | 12.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.7 | 16.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.6 | 3.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.6 | 5.7 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.6 | 4.0 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.6 | 10.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.6 | 2.8 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.5 | 2.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.5 | 9.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.5 | 34.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.5 | 2.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.5 | 9.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.4 | 3.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.4 | 1.3 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.4 | 2.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.4 | 7.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.4 | 6.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 13.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.4 | 5.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.4 | 3.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 11.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 9.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 2.4 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.3 | 23.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 7.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.3 | 4.0 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.3 | 8.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 1.8 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.3 | 3.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 8.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 24.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 2.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 9.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.3 | 33.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.3 | 2.5 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.3 | 3.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.3 | 8.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 3.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 4.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 11.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 1.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 2.0 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 12.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 2.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 20.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 4.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 0.9 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 2.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 3.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 0.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 5.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 6.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 2.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 24.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 7.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 1.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.4 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 4.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 13.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 23.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 40.8 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 14.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 4.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.8 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 3.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 64.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 7.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 0.7 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 1.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 2.2 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 20.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 2.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.8 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.1 | 0.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 18.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 20.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 6.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 1.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 5.9 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 17.9 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 3.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 4.1 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 1.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 52.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 1.6 | 67.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.8 | 29.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.7 | 109.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.7 | 43.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.7 | 22.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.6 | 133.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.6 | 40.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.6 | 21.8 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.5 | 67.4 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.5 | 24.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.5 | 5.9 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.4 | 6.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.4 | 32.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.3 | 3.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 10.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.3 | 14.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 7.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 6.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 9.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 6.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 3.3 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.2 | 3.9 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.2 | 9.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 14.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 6.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 2.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 8.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 5.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.7 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 4.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.8 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.6 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.5 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.8 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 237.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 2.1 | 26.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.9 | 43.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 1.2 | 27.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.1 | 13.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.0 | 21.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.9 | 24.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.9 | 16.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.9 | 32.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.7 | 17.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.7 | 15.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.7 | 21.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.6 | 10.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.6 | 12.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.5 | 39.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.5 | 6.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.5 | 19.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.5 | 12.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.4 | 19.5 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.4 | 12.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.4 | 5.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.4 | 5.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.4 | 4.0 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.4 | 9.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.4 | 23.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 4.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 16.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 22.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 12.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 9.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.3 | 6.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.3 | 42.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 11.3 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.2 | 6.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 2.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 19.9 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.2 | 8.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 5.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 3.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 4.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 7.1 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.2 | 5.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 3.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 4.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 2.0 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.1 | 4.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.9 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 7.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 13.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 7.8 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 1.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 27.8 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 9.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 3.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |