Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for MYB
Z-value: 9.05
Transcription factors associated with MYB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYB
|
ENSG00000118513.19 | MYB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYB | hg38_v1_chr6_+_135181268_135181322 | 0.33 | 7.5e-07 | Click! |
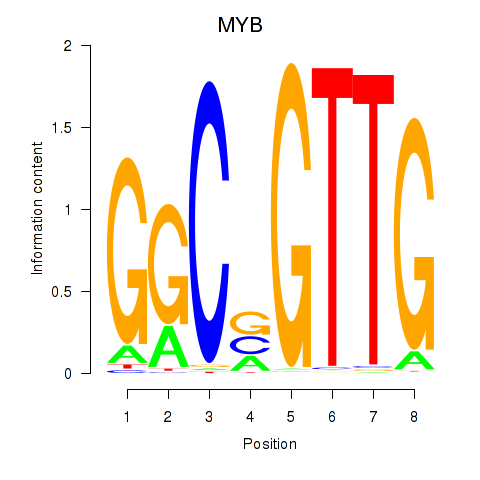
Activity profile of MYB motif
Sorted Z-values of MYB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYB
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 38.9 | 155.6 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 31.3 | 125.1 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 22.4 | 291.7 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 14.2 | 56.7 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 12.3 | 73.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 12.2 | 61.2 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 11.9 | 95.4 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 10.8 | 43.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 10.3 | 30.8 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 9.2 | 27.7 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 9.2 | 27.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 9.1 | 27.2 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 8.7 | 52.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 8.4 | 67.4 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 8.2 | 180.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 7.7 | 31.0 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 7.5 | 52.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 7.4 | 22.1 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 7.4 | 36.8 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 7.2 | 43.1 | GO:0015853 | adenine transport(GO:0015853) |
| 6.9 | 20.7 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 6.5 | 26.0 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 6.5 | 19.4 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 6.2 | 18.7 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 5.6 | 16.7 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 5.0 | 84.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 4.9 | 24.3 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 4.7 | 217.2 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 4.6 | 13.8 | GO:0015866 | ADP transport(GO:0015866) |
| 4.6 | 13.7 | GO:0072019 | proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 4.5 | 13.6 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 4.4 | 53.0 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 4.4 | 66.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 4.3 | 30.3 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 4.3 | 34.5 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 4.1 | 140.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 4.1 | 53.0 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 4.0 | 44.5 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 4.0 | 23.9 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 3.9 | 11.8 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 3.9 | 11.6 | GO:0051030 | snRNA transport(GO:0051030) |
| 3.8 | 19.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 3.8 | 22.9 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 3.8 | 11.4 | GO:0097698 | telomere maintenance via base-excision repair(GO:0097698) |
| 3.7 | 15.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 3.7 | 33.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 3.7 | 11.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 3.7 | 62.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 3.6 | 18.1 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 3.6 | 64.9 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 3.6 | 10.7 | GO:0034402 | mRNA export from nucleus in response to heat stress(GO:0031990) recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 3.6 | 10.7 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 3.4 | 54.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 3.4 | 376.0 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 3.4 | 3.4 | GO:1904884 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 3.3 | 101.8 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 3.2 | 6.5 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 3.2 | 9.7 | GO:0009386 | translational attenuation(GO:0009386) |
| 3.2 | 12.8 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 3.2 | 28.4 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 3.1 | 28.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 3.1 | 12.4 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 3.1 | 21.4 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 3.0 | 12.1 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 3.0 | 12.0 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 2.9 | 8.8 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) |
| 2.9 | 17.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 2.9 | 5.7 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 2.9 | 20.0 | GO:0060701 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 2.8 | 39.8 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 2.8 | 8.5 | GO:2000395 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) ubiquitin-dependent endocytosis(GO:0070086) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 2.8 | 14.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 2.8 | 5.6 | GO:2000349 | negative regulation of CD40 signaling pathway(GO:2000349) |
| 2.7 | 32.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 2.7 | 87.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 2.5 | 35.2 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 2.5 | 82.3 | GO:0071174 | mitotic spindle checkpoint(GO:0071174) |
| 2.5 | 24.7 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 2.5 | 27.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 2.4 | 21.5 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 2.4 | 28.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 2.4 | 18.9 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 2.3 | 9.3 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 2.3 | 18.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 2.3 | 6.9 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 2.3 | 13.7 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 2.3 | 6.9 | GO:2000847 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 2.2 | 8.9 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 2.2 | 160.8 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 2.2 | 17.5 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 2.1 | 27.7 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 2.1 | 8.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 2.1 | 14.6 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 2.1 | 8.3 | GO:0018158 | protein oxidation(GO:0018158) |
| 2.1 | 20.8 | GO:0019695 | choline metabolic process(GO:0019695) |
| 2.1 | 10.3 | GO:0030242 | pexophagy(GO:0030242) |
| 2.0 | 12.0 | GO:0044211 | CTP salvage(GO:0044211) |
| 2.0 | 5.9 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 2.0 | 43.4 | GO:1902001 | carnitine shuttle(GO:0006853) fatty acid transmembrane transport(GO:1902001) |
| 2.0 | 21.7 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 2.0 | 45.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 2.0 | 29.4 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 2.0 | 37.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 1.9 | 5.8 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 1.9 | 15.5 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 1.9 | 5.7 | GO:0033242 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) regulation of cellular amino acid biosynthetic process(GO:2000282) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 1.9 | 13.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 1.9 | 43.3 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 1.9 | 22.4 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 1.9 | 11.2 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 1.8 | 16.5 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 1.8 | 14.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 1.8 | 15.9 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 1.8 | 12.4 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 1.8 | 8.8 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 1.7 | 87.0 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 1.7 | 13.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 1.7 | 15.6 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 1.7 | 107.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 1.7 | 11.7 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 1.7 | 8.3 | GO:0010868 | negative regulation of triglyceride biosynthetic process(GO:0010868) |
| 1.7 | 18.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 1.6 | 6.5 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 1.6 | 11.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 1.6 | 6.4 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 1.6 | 12.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.6 | 4.7 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 1.5 | 6.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 1.5 | 44.2 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 1.5 | 3.0 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 1.5 | 13.7 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 1.5 | 4.6 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 1.5 | 4.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 1.5 | 27.6 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 1.4 | 21.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 1.4 | 32.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 1.4 | 18.1 | GO:0045008 | depyrimidination(GO:0045008) |
| 1.4 | 19.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.4 | 16.6 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 1.3 | 10.7 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 1.3 | 4.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 1.3 | 35.7 | GO:0072662 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 1.3 | 15.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 1.3 | 25.0 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 1.3 | 10.5 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 1.3 | 3.9 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 1.3 | 24.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 1.3 | 7.7 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 1.3 | 9.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.3 | 10.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 1.3 | 44.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 1.3 | 7.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 1.3 | 8.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 1.3 | 20.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 1.2 | 6.2 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 1.2 | 4.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.2 | 51.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 1.2 | 4.8 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 1.2 | 107.6 | GO:0070671 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 1.2 | 6.9 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 1.1 | 11.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 1.1 | 29.7 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 1.1 | 13.5 | GO:0098969 | neurotransmitter receptor transport to plasma membrane(GO:0098877) neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) protein localization to postsynaptic membrane(GO:1903539) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 1.1 | 17.9 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 1.1 | 14.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 1.1 | 5.3 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 1.1 | 3.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 1.0 | 27.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 1.0 | 37.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 1.0 | 5.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 1.0 | 10.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 1.0 | 75.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 1.0 | 23.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 1.0 | 4.8 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.9 | 5.6 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.9 | 58.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.9 | 5.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.9 | 14.0 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.9 | 84.7 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.9 | 10.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.9 | 10.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.8 | 1.7 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.8 | 10.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.8 | 7.5 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.8 | 70.6 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.8 | 6.5 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.8 | 2.4 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.8 | 12.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.8 | 7.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.8 | 3.1 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.8 | 3.9 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.8 | 11.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.7 | 44.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.7 | 17.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.7 | 3.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.7 | 104.4 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.7 | 2.7 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.7 | 6.7 | GO:0030656 | regulation of vitamin metabolic process(GO:0030656) |
| 0.7 | 8.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.7 | 2.6 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.7 | 5.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.6 | 10.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.6 | 3.2 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.6 | 3.8 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.6 | 1.8 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.6 | 8.9 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.6 | 10.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.6 | 15.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.5 | 4.8 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.5 | 2.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.5 | 25.8 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.5 | 5.7 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.5 | 9.8 | GO:0000732 | strand displacement(GO:0000732) |
| 0.5 | 7.7 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.5 | 3.0 | GO:0046719 | regulation by virus of viral protein levels in host cell(GO:0046719) |
| 0.5 | 8.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.5 | 16.6 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.5 | 5.8 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.5 | 4.8 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.5 | 1.9 | GO:0010454 | negative regulation of cell fate commitment(GO:0010454) |
| 0.5 | 2.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.5 | 6.9 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.5 | 1.8 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.5 | 10.0 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.4 | 28.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.4 | 62.9 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.4 | 4.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.4 | 2.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.4 | 18.1 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.4 | 10.5 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.4 | 1.2 | GO:0051365 | leading edge cell differentiation(GO:0035026) cellular response to potassium ion starvation(GO:0051365) |
| 0.4 | 5.8 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.4 | 4.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.4 | 6.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.4 | 5.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.4 | 1.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.3 | 2.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 10.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.3 | 1.0 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.3 | 6.8 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.3 | 3.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.3 | 23.4 | GO:0048208 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.3 | 6.1 | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.3 | 3.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.3 | 11.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.3 | 4.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.3 | 3.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.3 | 7.0 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.3 | 1.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.3 | 1.1 | GO:1901723 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) visceral serous pericardium development(GO:0061032) S-shaped body morphogenesis(GO:0072050) negative regulation of glomerular mesangial cell proliferation(GO:0072125) posterior mesonephric tubule development(GO:0072166) metanephric S-shaped body morphogenesis(GO:0072284) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) negative regulation of glomerulus development(GO:0090194) negative regulation of cell proliferation involved in kidney development(GO:1901723) |
| 0.3 | 1.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.3 | 41.0 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.2 | 23.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 9.9 | GO:1990090 | cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.2 | 1.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.2 | 1.7 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.2 | 4.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.2 | 10.7 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 6.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 68.0 | GO:0030335 | positive regulation of cell migration(GO:0030335) |
| 0.2 | 18.9 | GO:0006413 | translational initiation(GO:0006413) |
| 0.2 | 0.9 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 2.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 3.0 | GO:1901984 | negative regulation of protein acetylation(GO:1901984) |
| 0.2 | 11.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.2 | 0.5 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.2 | 7.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 4.3 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 13.5 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.1 | 9.0 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 4.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 3.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 2.9 | GO:0017145 | stem cell division(GO:0017145) |
| 0.1 | 14.4 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.9 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.8 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 8.4 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.1 | 2.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 4.7 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.1 | 3.3 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 2.1 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.6 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 2.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 3.0 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 2.7 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 5.0 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 2.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 1.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 2.8 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.3 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 12.7 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 2.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 2.6 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 1.1 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 2.7 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 6.1 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.4 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.7 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.3 | 256.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 12.5 | 125.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 11.2 | 280.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 9.4 | 151.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 9.0 | 80.6 | GO:0001520 | outer dense fiber(GO:0001520) |
| 8.6 | 34.6 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 8.5 | 25.5 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 6.9 | 20.7 | GO:0055087 | Ski complex(GO:0055087) |
| 6.4 | 19.3 | GO:1990393 | 3M complex(GO:1990393) |
| 6.4 | 70.0 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 6.2 | 18.6 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 6.1 | 54.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 5.9 | 53.0 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 5.6 | 28.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 5.3 | 26.5 | GO:0000796 | condensin complex(GO:0000796) |
| 5.3 | 26.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 5.3 | 100.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 5.2 | 15.6 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 5.0 | 40.4 | GO:0000243 | commitment complex(GO:0000243) |
| 5.0 | 75.1 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 4.9 | 141.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 4.9 | 14.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 4.8 | 58.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 4.6 | 55.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 4.5 | 300.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 4.4 | 31.0 | GO:0031415 | NatA complex(GO:0031415) |
| 4.3 | 12.8 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 4.3 | 17.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 3.9 | 11.8 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 3.8 | 19.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 3.5 | 72.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 3.3 | 39.8 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 3.2 | 51.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 3.2 | 12.8 | GO:0071817 | MMXD complex(GO:0071817) |
| 3.0 | 30.4 | GO:0043203 | axon hillock(GO:0043203) |
| 3.0 | 18.1 | GO:0032279 | axonemal microtubule(GO:0005879) asymmetric synapse(GO:0032279) |
| 2.9 | 32.1 | GO:0032039 | integrator complex(GO:0032039) |
| 2.9 | 22.9 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 2.8 | 19.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 2.8 | 8.3 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 2.7 | 10.7 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 2.7 | 13.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 2.7 | 8.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 2.6 | 62.9 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 2.6 | 12.8 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 2.5 | 22.4 | GO:0000346 | transcription export complex(GO:0000346) |
| 2.5 | 9.8 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 2.5 | 19.6 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 2.4 | 48.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 2.1 | 8.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 2.1 | 10.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 2.0 | 20.5 | GO:0008278 | cohesin complex(GO:0008278) |
| 2.0 | 218.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 1.9 | 46.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 1.9 | 43.1 | GO:0046930 | pore complex(GO:0046930) |
| 1.8 | 29.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 1.8 | 12.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 1.8 | 22.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.7 | 8.7 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.7 | 6.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.6 | 4.9 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.6 | 4.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.6 | 19.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 1.6 | 83.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 1.5 | 7.7 | GO:0016589 | NURF complex(GO:0016589) |
| 1.5 | 13.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 1.5 | 16.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 1.5 | 22.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 1.4 | 48.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 1.4 | 17.0 | GO:0071203 | F-actin capping protein complex(GO:0008290) WASH complex(GO:0071203) |
| 1.4 | 41.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 1.4 | 12.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 1.4 | 12.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 1.4 | 12.4 | GO:0000938 | GARP complex(GO:0000938) |
| 1.4 | 56.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 1.3 | 147.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 1.3 | 62.7 | GO:0005844 | polysome(GO:0005844) |
| 1.3 | 6.5 | GO:0033503 | HULC complex(GO:0033503) |
| 1.3 | 50.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 1.3 | 3.8 | GO:0044393 | microspike(GO:0044393) |
| 1.2 | 15.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex III(GO:0045275) |
| 1.2 | 43.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 1.1 | 4.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.1 | 10.8 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 1.0 | 11.3 | GO:0005638 | lamin filament(GO:0005638) |
| 1.0 | 10.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 1.0 | 6.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 1.0 | 98.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 1.0 | 4.0 | GO:0071942 | XPC complex(GO:0071942) |
| 1.0 | 13.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 1.0 | 12.0 | GO:0034709 | methylosome(GO:0034709) |
| 1.0 | 61.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 1.0 | 27.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 1.0 | 12.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.9 | 10.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.9 | 8.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.9 | 24.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.9 | 77.4 | GO:0031672 | A band(GO:0031672) |
| 0.8 | 2.5 | GO:0071754 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.8 | 102.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.8 | 10.0 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.7 | 4.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.7 | 28.6 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.7 | 6.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.7 | 59.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.7 | 13.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.7 | 63.6 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.7 | 50.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.6 | 9.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.5 | 13.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.5 | 22.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.5 | 11.1 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.5 | 4.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.4 | 10.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.4 | 3.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 51.2 | GO:0031514 | motile cilium(GO:0031514) |
| 0.4 | 40.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 143.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.4 | 5.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.4 | 11.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.4 | 5.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 77.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.4 | 1.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 4.1 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.3 | 32.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.3 | 2.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.3 | 52.9 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.3 | 6.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 6.7 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.3 | 45.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.3 | 1.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 27.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.3 | 0.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 8.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 6.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 3.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 12.9 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.2 | 8.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 7.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 20.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 14.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 1.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 21.0 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.2 | 12.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 5.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 11.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 7.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 10.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 6.8 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 27.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 2.3 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.1 | 44.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 3.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 15.8 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 5.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 12.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 30.8 | GO:1990904 | ribonucleoprotein complex(GO:1990904) |
| 0.1 | 1.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 5.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 64.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 1.7 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 1.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 177.5 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.9 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 7.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 1.8 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 2.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.2 | GO:0031901 | early endosome membrane(GO:0031901) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 25.0 | 125.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 20.4 | 101.8 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 18.4 | 128.8 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 17.5 | 70.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 15.6 | 62.2 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) |
| 14.6 | 321.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 14.0 | 83.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 13.0 | 155.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 12.6 | 63.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 10.7 | 74.9 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 8.7 | 43.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 8.1 | 48.9 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 8.1 | 40.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 8.0 | 15.9 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 7.9 | 39.7 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 7.5 | 52.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 7.2 | 21.7 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 6.1 | 18.3 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 6.1 | 122.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 5.8 | 58.0 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 5.8 | 34.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 5.4 | 43.1 | GO:0015288 | porin activity(GO:0015288) |
| 5.2 | 20.8 | GO:0033265 | choline binding(GO:0033265) |
| 5.1 | 293.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 4.7 | 37.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 4.6 | 64.9 | GO:0031386 | protein tag(GO:0031386) |
| 4.5 | 13.6 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 4.2 | 34.0 | GO:0050733 | RS domain binding(GO:0050733) |
| 4.1 | 53.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 3.7 | 22.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 3.7 | 55.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 3.6 | 18.1 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 3.6 | 21.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 3.4 | 13.7 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 3.4 | 10.2 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 3.3 | 39.8 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 3.3 | 13.1 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 3.3 | 29.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 3.3 | 22.9 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 3.2 | 72.7 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 3.1 | 118.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 3.0 | 12.0 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 2.9 | 134.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 2.9 | 20.0 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 2.9 | 28.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 2.8 | 11.4 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 2.8 | 25.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 2.8 | 123.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 2.7 | 10.7 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 2.7 | 10.7 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 2.6 | 12.8 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 2.4 | 21.5 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 2.4 | 11.8 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 2.3 | 7.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 2.3 | 7.0 | GO:0000252 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) sterol-4-alpha-carboxylate 3-dehydrogenase (decarboxylating) activity(GO:0047012) |
| 2.3 | 21.0 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) |
| 2.3 | 20.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 2.2 | 13.5 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 2.2 | 31.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 2.2 | 56.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 2.1 | 23.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 2.1 | 8.3 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 2.0 | 14.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 2.0 | 31.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 2.0 | 15.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 2.0 | 13.8 | GO:0005347 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 1.9 | 5.7 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 1.9 | 13.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 1.7 | 33.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 1.7 | 12.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 1.7 | 30.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 1.7 | 11.6 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 1.6 | 65.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 1.6 | 22.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.4 | 10.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 1.4 | 51.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 1.4 | 39.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 1.4 | 9.6 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 1.4 | 83.2 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 1.3 | 24.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 1.3 | 16.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 1.3 | 8.9 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 1.2 | 11.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 1.2 | 8.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 1.2 | 4.7 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 1.1 | 44.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 1.1 | 36.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 1.1 | 3.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 1.0 | 13.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 1.0 | 78.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 1.0 | 11.0 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 1.0 | 9.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 1.0 | 14.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 1.0 | 17.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 1.0 | 20.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 1.0 | 20.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 1.0 | 4.8 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.9 | 17.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.9 | 12.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.9 | 66.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.9 | 17.6 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.9 | 73.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.8 | 6.7 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.8 | 20.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.8 | 4.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.8 | 14.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.7 | 4.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.7 | 139.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.6 | 50.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.6 | 3.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.6 | 37.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.6 | 11.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.6 | 26.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.6 | 10.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.6 | 15.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.6 | 8.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.6 | 2.3 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.6 | 13.0 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.6 | 6.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.6 | 20.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.5 | 6.5 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.5 | 3.8 | GO:0097617 | annealing helicase activity(GO:0036310) annealing activity(GO:0097617) |
| 0.5 | 2.6 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.5 | 2.6 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.5 | 2.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.5 | 25.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.5 | 8.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.5 | 2.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.5 | 34.6 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.4 | 4.0 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.4 | 5.8 | GO:0016018 | cyclosporin A binding(GO:0016018) virion binding(GO:0046790) |
| 0.4 | 4.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 2.6 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.4 | 62.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.4 | 14.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.4 | 10.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.4 | 15.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.4 | 15.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.4 | 5.8 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.4 | 11.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.4 | 1.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.4 | 3.4 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.4 | 10.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.4 | 82.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.4 | 13.7 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.4 | 42.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.4 | 5.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 33.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.3 | 10.0 | GO:0015149 | hexose transmembrane transporter activity(GO:0015149) |
| 0.3 | 10.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.3 | 43.4 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.3 | 1.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 1.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.3 | 22.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.3 | 22.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.3 | 5.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.3 | 4.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.3 | 22.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.3 | 1.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 13.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 2.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 5.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 1.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 259.5 | GO:0003723 | RNA binding(GO:0003723) |
| 0.2 | 12.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 20.8 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.2 | 8.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 16.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.2 | 2.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 45.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.2 | 7.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 38.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.2 | 4.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 2.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 2.7 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.1 | 4.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 9.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.6 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 46.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 2.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.5 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 2.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 2.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 2.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 2.0 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 1.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 1.0 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 318.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 3.9 | 154.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 2.9 | 142.6 | PID MYC PATHWAY | C-MYC pathway |
| 2.3 | 136.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 2.0 | 58.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 1.9 | 166.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 1.8 | 86.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 1.8 | 59.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 1.8 | 62.1 | PID ATM PATHWAY | ATM pathway |
| 1.6 | 27.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 1.2 | 45.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 1.2 | 55.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 1.0 | 66.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.8 | 22.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.7 | 21.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.7 | 51.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.7 | 37.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.7 | 58.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.7 | 23.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.7 | 48.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.6 | 18.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.6 | 7.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.5 | 38.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.5 | 8.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.4 | 7.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.3 | 27.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 7.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 13.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 8.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.2 | 6.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 9.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 4.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 11.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.2 | 8.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 5.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 2.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 6.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 5.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 6.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 3.8 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 1.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 1.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.1 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 304.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 5.3 | 175.8 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 4.9 | 73.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 4.3 | 68.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 3.9 | 27.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 3.5 | 133.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 3.3 | 30.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 3.1 | 18.7 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 2.8 | 5.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 2.7 | 70.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 2.2 | 95.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 2.0 | 180.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 1.7 | 141.9 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 1.5 | 29.4 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 1.4 | 52.1 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 1.3 | 21.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 1.3 | 27.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 1.3 | 20.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 1.3 | 23.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 1.2 | 14.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.2 | 8.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 1.2 | 26.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 1.2 | 46.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 1.1 | 18.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.1 | 40.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 1.0 | 32.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 1.0 | 111.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 1.0 | 51.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 1.0 | 27.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 1.0 | 5.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 1.0 | 44.9 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.9 | 3.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.9 | 157.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.9 | 18.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.9 | 17.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.8 | 27.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.8 | 48.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.8 | 22.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.8 | 10.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.7 | 64.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.7 | 10.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.7 | 39.1 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.7 | 42.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 11.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.6 | 10.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.6 | 7.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.5 | 1.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.5 | 8.2 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.5 | 11.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 75.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.4 | 14.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.4 | 13.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.4 | 18.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.4 | 52.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.4 | 6.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.4 | 14.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.4 | 5.7 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.3 | 10.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.3 | 9.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.3 | 6.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 16.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 1.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.3 | 12.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 4.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 16.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 5.8 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.2 | 11.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 5.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 4.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 5.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 5.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 2.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.1 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.5 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.9 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |