Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
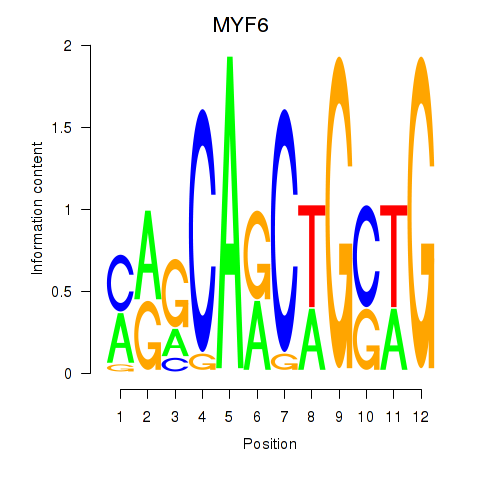
Results for MYF6
Z-value: 3.30
Transcription factors associated with MYF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYF6
|
ENSG00000111046.4 | MYF6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYF6 | hg38_v1_chr12_+_80707625_80707642 | 0.25 | 1.4e-04 | Click! |
Activity profile of MYF6 motif
Sorted Z-values of MYF6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYF6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 22.6 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 4.6 | 13.8 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 4.2 | 12.7 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 4.1 | 12.3 | GO:0001794 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 4.1 | 20.3 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 4.0 | 12.1 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 3.3 | 9.8 | GO:2000374 | lysosomal membrane organization(GO:0097212) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 3.1 | 3.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 3.0 | 9.0 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 2.8 | 11.3 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 2.5 | 17.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 2.4 | 7.2 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 2.3 | 7.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 2.3 | 6.8 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 2.3 | 13.6 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 2.2 | 11.0 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 2.2 | 8.7 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 2.1 | 12.7 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 2.1 | 25.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 2.0 | 5.9 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 1.9 | 19.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.9 | 5.8 | GO:0016476 | calcium-dependent cell-matrix adhesion(GO:0016340) regulation of embryonic cell shape(GO:0016476) |
| 1.9 | 30.4 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 1.9 | 32.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.9 | 3.7 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 1.9 | 1.9 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 1.9 | 7.4 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 1.7 | 8.3 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.7 | 5.0 | GO:0060596 | mammary placode formation(GO:0060596) His-Purkinje system cell differentiation(GO:0060932) |
| 1.6 | 4.9 | GO:0043449 | olfactory learning(GO:0008355) cellular alkene metabolic process(GO:0043449) |
| 1.6 | 4.8 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 1.6 | 4.7 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 1.5 | 9.3 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 1.5 | 6.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 1.5 | 4.6 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 1.5 | 4.5 | GO:1904437 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 1.4 | 14.4 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 1.4 | 7.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 1.4 | 4.2 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 1.4 | 9.7 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 1.4 | 4.1 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 1.4 | 5.5 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 1.3 | 5.3 | GO:0071879 | UDP-glucose catabolic process(GO:0006258) positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 1.3 | 6.6 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 1.3 | 5.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 1.3 | 3.8 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 1.2 | 24.7 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 1.2 | 18.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.2 | 15.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 1.2 | 4.7 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 1.2 | 8.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 1.2 | 3.5 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 1.1 | 4.5 | GO:1990834 | response to odorant(GO:1990834) |
| 1.1 | 2.2 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 1.1 | 4.3 | GO:1903891 | regulation of ATF6-mediated unfolded protein response(GO:1903891) |
| 1.1 | 5.4 | GO:0001555 | oocyte growth(GO:0001555) regulation of progesterone secretion(GO:2000870) |
| 1.1 | 15.0 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 1.0 | 3.1 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 1.0 | 2.0 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 1.0 | 4.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 1.0 | 11.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 1.0 | 4.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 1.0 | 4.0 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.0 | 4.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.0 | 10.8 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 1.0 | 4.9 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 1.0 | 7.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.9 | 6.5 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.9 | 2.8 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.9 | 7.4 | GO:1902473 | regulation of protein localization to synapse(GO:1902473) |
| 0.9 | 8.3 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.9 | 2.7 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.9 | 4.6 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.9 | 71.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.9 | 8.7 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.9 | 2.6 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.8 | 7.4 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.8 | 2.4 | GO:0061163 | endoplasmic reticulum polarization(GO:0061163) actin filament bundle retrograde transport(GO:0061573) actin filament bundle distribution(GO:0070650) |
| 0.8 | 8.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.8 | 3.9 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.8 | 14.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.8 | 7.7 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.8 | 4.5 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.7 | 3.7 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.7 | 3.7 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.7 | 2.9 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.7 | 2.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.7 | 9.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.7 | 5.6 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.7 | 2.8 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.7 | 2.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.7 | 1.3 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.6 | 2.6 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.6 | 2.6 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.6 | 12.4 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.6 | 2.4 | GO:1902938 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.6 | 3.5 | GO:0036100 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.6 | 2.3 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.6 | 4.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.6 | 6.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.5 | 1.6 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.5 | 2.1 | GO:0019086 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.5 | 4.8 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.5 | 1.6 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.5 | 4.1 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.5 | 5.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.5 | 5.0 | GO:0046959 | habituation(GO:0046959) |
| 0.5 | 3.4 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.5 | 3.4 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.5 | 1.0 | GO:0061047 | regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.5 | 4.3 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.5 | 1.4 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.5 | 3.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.5 | 3.7 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.5 | 1.4 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.4 | 3.6 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.4 | 1.8 | GO:0097491 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.4 | 4.8 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.4 | 1.3 | GO:0034699 | response to luteinizing hormone(GO:0034699) |
| 0.4 | 4.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 9.1 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.4 | 6.9 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.4 | 1.3 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.4 | 2.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.4 | 1.7 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.4 | 3.8 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.4 | 0.8 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.4 | 2.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.4 | 3.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.4 | 4.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.4 | 3.5 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.4 | 8.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.4 | 1.9 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.4 | 4.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 | 18.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.4 | 2.6 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.4 | 3.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.4 | 5.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.4 | 1.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.4 | 1.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.4 | 2.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.3 | 1.0 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) |
| 0.3 | 4.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.3 | 4.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.3 | 1.7 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.3 | 4.4 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.3 | 4.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 2.3 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.3 | 1.9 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.3 | 1.6 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.3 | 2.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 | 7.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.3 | 0.9 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.3 | 0.9 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.3 | 5.7 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.3 | 3.5 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.3 | 3.5 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.3 | 3.2 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.3 | 1.4 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 1.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.3 | 5.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.3 | 0.8 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.3 | 3.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.3 | 1.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.3 | 5.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.3 | 3.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.2 | 3.7 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.2 | 0.5 | GO:0051462 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) positive regulation of cortisol secretion(GO:0051464) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.2 | 2.4 | GO:0003360 | brainstem development(GO:0003360) |
| 0.2 | 1.4 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 | 1.9 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 1.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.2 | 0.7 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.2 | 8.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 1.6 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.2 | 3.2 | GO:0042659 | spinal cord association neuron differentiation(GO:0021527) regulation of cell fate specification(GO:0042659) |
| 0.2 | 1.1 | GO:0003068 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 2.0 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.2 | 2.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 2.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.2 | 16.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 2.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 3.9 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.2 | 5.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 6.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 0.8 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.2 | 2.9 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.2 | 2.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 8.6 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.2 | 17.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 5.6 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.2 | 2.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 6.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 1.0 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.2 | 1.2 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.2 | 1.0 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.2 | 0.6 | GO:0031587 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.2 | 1.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 0.6 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.2 | 0.8 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.2 | 0.6 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 3.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 1.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 1.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 6.5 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.2 | 25.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 0.5 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.2 | 0.9 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.2 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.2 | 1.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.2 | 4.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 0.7 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.2 | 1.3 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.2 | 5.1 | GO:0021988 | olfactory lobe development(GO:0021988) |
| 0.2 | 1.6 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.2 | 2.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.2 | 1.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 1.7 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.2 | 1.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.2 | 1.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.2 | 1.5 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 0.6 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D3 metabolic process(GO:0070640) |
| 0.1 | 0.6 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 | 4.6 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.1 | 6.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 1.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 2.5 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 3.9 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 4.8 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 4.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 1.7 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.1 | 3.6 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 3.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.8 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 1.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 1.1 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.1 | 1.0 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 2.2 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.1 | 0.7 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 4.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 2.6 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.1 | 4.4 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.1 | 0.2 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 2.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.4 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.8 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 1.3 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 1.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 1.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 2.7 | GO:0033866 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.1 | 3.9 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 1.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 3.9 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 3.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.4 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 1.0 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.6 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.1 | 3.8 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 1.0 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.7 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.8 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.5 | GO:0014061 | regulation of norepinephrine secretion(GO:0014061) |
| 0.1 | 1.6 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 0.4 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.4 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 14.1 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 0.3 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.4 | GO:0048619 | hindgut morphogenesis(GO:0007442) embryonic hindgut morphogenesis(GO:0048619) hindgut development(GO:0061525) |
| 0.1 | 0.3 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.1 | 1.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 1.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) inner dynein arm assembly(GO:0036159) |
| 0.1 | 5.4 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 0.6 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.1 | 1.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.7 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.5 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 1.0 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.1 | 1.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 1.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 1.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.1 | 3.0 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.1 | 2.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 2.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 2.7 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 1.5 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.1 | 1.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 2.0 | GO:0050779 | RNA destabilization(GO:0050779) |
| 0.1 | 0.6 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.1 | 1.2 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 4.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 4.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 2.3 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.1 | 0.7 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 2.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 4.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 4.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 0.8 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.1 | 0.2 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.1 | 7.4 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.0 | 0.5 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 3.6 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.5 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 1.1 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.7 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.6 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.0 | 1.0 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 1.6 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.4 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) |
| 0.0 | 1.6 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 1.0 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.5 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.6 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.1 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 7.7 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.6 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 1.6 | GO:0008585 | female gonad development(GO:0008585) |
| 0.0 | 0.3 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 1.4 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 2.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.4 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 1.5 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 1.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 1.1 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.0 | 2.9 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 3.2 | GO:0072659 | protein localization to plasma membrane(GO:0072659) |
| 0.0 | 0.7 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 2.9 | GO:0007015 | actin filament organization(GO:0007015) |
| 0.0 | 2.0 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.5 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 1.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 13.9 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 3.2 | 9.7 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 2.4 | 38.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 2.3 | 7.0 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 2.2 | 17.6 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 1.8 | 11.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 1.7 | 5.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 1.5 | 4.6 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 1.4 | 16.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.2 | 10.0 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 1.2 | 15.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.0 | 14.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.0 | 10.0 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.9 | 5.6 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.9 | 23.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.9 | 9.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.9 | 10.8 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.9 | 4.5 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.8 | 5.0 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.8 | 8.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.8 | 10.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.8 | 4.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.7 | 3.7 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.6 | 12.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.6 | 7.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.6 | 7.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.6 | 3.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.6 | 13.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.5 | 4.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.5 | 4.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.5 | 1.6 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.5 | 32.3 | GO:0005844 | polysome(GO:0005844) |
| 0.5 | 1.5 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.4 | 3.6 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.4 | 6.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.4 | 6.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.4 | 3.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 12.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.4 | 5.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 1.9 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.4 | 7.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.3 | 2.4 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.3 | 2.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 3.6 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 2.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 12.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.3 | 5.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 1.3 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.3 | 3.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 4.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 17.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 0.9 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 24.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 2.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.3 | 5.6 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.3 | 12.6 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 2.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.3 | 21.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 8.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 2.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 3.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 4.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 1.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 5.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 2.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 0.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 20.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 0.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 1.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 7.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 1.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 0.9 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 3.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 2.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 1.0 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 1.0 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 1.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 1.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 11.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.2 | 1.9 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 21.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 37.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 0.9 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 5.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 2.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 2.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 14.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 10.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 2.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 2.3 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 24.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 30.1 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.1 | 0.6 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 2.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 32.8 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 8.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 11.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 1.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 4.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 4.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 1.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 3.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 2.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 4.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 5.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 10.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 1.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 4.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 5.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 104.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.6 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.7 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 0.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 7.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 84.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 0.9 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 7.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 9.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 9.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 1.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.5 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.0 | 4.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.6 | GO:0044439 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.2 | GO:0030120 | vesicle coat(GO:0030120) |
| 0.0 | 7.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.8 | GO:0045202 | synapse(GO:0045202) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 20.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 4.2 | 25.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 3.5 | 13.9 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 2.8 | 11.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 2.7 | 13.6 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 2.4 | 9.7 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 2.4 | 7.2 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 2.1 | 12.7 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 1.8 | 9.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 1.7 | 10.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 1.7 | 6.8 | GO:0032810 | sterol response element binding(GO:0032810) |
| 1.6 | 8.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 1.4 | 5.8 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 1.4 | 18.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.4 | 29.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.4 | 4.1 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 1.3 | 17.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.3 | 4.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 1.3 | 6.6 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 1.3 | 38.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.2 | 5.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.2 | 11.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 1.2 | 3.7 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 1.2 | 76.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 1.2 | 3.6 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 1.2 | 3.5 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 1.1 | 5.6 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 1.1 | 8.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 1.1 | 4.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 1.1 | 5.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.0 | 3.1 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 1.0 | 12.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.9 | 8.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.9 | 7.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.9 | 9.8 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.9 | 3.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.9 | 4.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.9 | 2.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.8 | 5.6 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.8 | 3.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.8 | 7.0 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.8 | 3.8 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.7 | 3.7 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.7 | 5.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.7 | 2.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.7 | 3.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.7 | 5.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.7 | 4.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.7 | 3.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.7 | 4.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.7 | 6.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.7 | 10.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.6 | 3.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.6 | 35.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.6 | 12.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.5 | 4.9 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.5 | 1.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.5 | 2.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.5 | 1.6 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.5 | 2.7 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.5 | 2.0 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.5 | 4.5 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.5 | 7.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.5 | 5.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.5 | 1.4 | GO:0036505 | prosaposin receptor activity(GO:0036505) |
| 0.5 | 5.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.5 | 4.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.5 | 12.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.5 | 4.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.4 | 1.3 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.4 | 4.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.4 | 1.3 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.4 | 14.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.4 | 3.0 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.4 | 1.7 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.4 | 9.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 2.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.4 | 4.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 1.7 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.4 | 6.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.4 | 14.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.4 | 3.7 | GO:0043426 | MRF binding(GO:0043426) |
| 0.4 | 2.4 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.4 | 2.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.4 | 3.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.4 | 5.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.4 | 22.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 2.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.3 | 1.7 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.3 | 4.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 4.0 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.3 | 8.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.3 | 2.3 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.3 | 5.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 5.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 1.8 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.3 | 5.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 4.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 2.6 | GO:0089720 | caspase binding(GO:0089720) |
| 0.3 | 34.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 2.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.3 | 1.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 2.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 4.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 5.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.3 | 0.8 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.3 | 4.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.3 | 4.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.3 | 1.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 2.0 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.3 | 3.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 1.0 | GO:0035473 | lipase binding(GO:0035473) |
| 0.2 | 1.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 2.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 1.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 7.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 7.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 2.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 1.9 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 2.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 11.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 1.8 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.2 | 4.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 1.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 4.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 0.8 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 1.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 2.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 2.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 29.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 6.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 8.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 1.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 0.5 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.2 | 14.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 2.6 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 0.5 | GO:0009931 | calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 1.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 0.5 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 3.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 0.6 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 1.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 1.9 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 1.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 0.8 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 1.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.8 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 1.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.2 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.1 | 2.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 4.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 15.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 13.6 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) |
| 0.1 | 0.8 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.3 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.1 | 5.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 4.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 10.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 3.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 2.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 7.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 2.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 5.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 9.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 0.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.6 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.1 | 1.0 | GO:0008432 | MAP-kinase scaffold activity(GO:0005078) JUN kinase binding(GO:0008432) |
| 0.1 | 2.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.8 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 0.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 1.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 7.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.6 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 9.6 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 0.9 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 8.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 7.6 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 6.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 4.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 11.1 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.1 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.6 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 27.5 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 1.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 6.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 3.1 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0034437 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 3.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 2.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 9.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.6 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 1.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 1.3 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 3.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0035586 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) purinergic receptor activity(GO:0035586) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 7.9 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 2.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 42.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.8 | 20.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.8 | 43.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.6 | 21.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.6 | 11.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.5 | 19.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.4 | 77.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.4 | 11.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 17.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.3 | 9.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 5.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 1.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.2 | 5.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 0.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 4.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 3.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 14.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 3.9 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 1.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 11.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 0.8 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 1.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 2.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 3.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 40.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 5.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 11.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 13.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 17.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 10.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 24.5 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.1 | 2.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 12.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 1.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 4.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 2.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 0.6 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 2.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 59.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.8 | 20.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.7 | 13.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.7 | 22.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.7 | 4.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.7 | 18.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.6 | 12.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.6 | 38.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.5 | 17.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.5 | 9.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.4 | 5.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.4 | 9.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 9.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 4.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 3.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.3 | 13.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 8.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 7.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 10.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.3 | 4.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 10.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 3.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 32.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 23.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 0.9 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.2 | 4.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 8.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 5.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 4.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 5.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 8.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 6.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 5.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.2 | 2.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 1.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 12.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 5.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 2.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 13.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 9.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 7.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 5.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 6.3 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 4.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.9 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 4.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.7 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 4.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.4 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 1.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 1.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 2.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 4.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 0.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.2 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.0 | 3.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 1.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 3.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 2.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.4 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |