Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
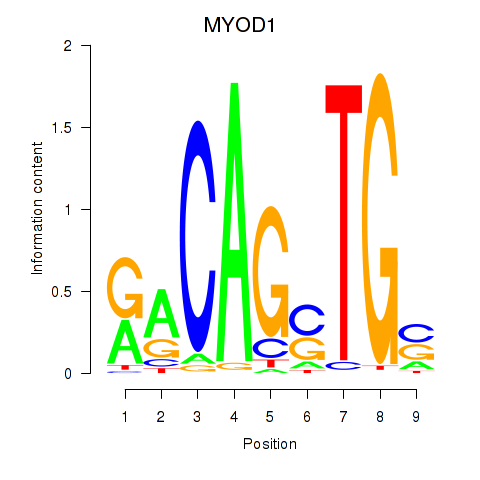
Results for MYOD1
Z-value: 7.02
Transcription factors associated with MYOD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYOD1
|
ENSG00000129152.4 | MYOD1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYOD1 | hg38_v1_chr11_+_17719564_17719577 | 0.54 | 1.2e-17 | Click! |
Activity profile of MYOD1 motif
Sorted Z-values of MYOD1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYOD1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.6 | 87.6 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 12.5 | 37.5 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 6.0 | 17.9 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 5.7 | 22.9 | GO:0090299 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 5.0 | 20.2 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 4.8 | 38.2 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 4.4 | 17.6 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 4.3 | 17.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 3.5 | 10.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 3.3 | 9.8 | GO:0048769 | skeletal muscle myosin thick filament assembly(GO:0030241) sarcomerogenesis(GO:0048769) |
| 3.1 | 12.2 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 3.0 | 27.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 2.8 | 22.8 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 2.7 | 13.3 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 2.6 | 20.8 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 2.5 | 7.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 2.5 | 9.8 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 2.4 | 7.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 2.4 | 9.4 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 2.3 | 20.9 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 2.1 | 137.0 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 2.1 | 14.5 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 2.0 | 8.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 2.0 | 6.0 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 2.0 | 10.0 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 2.0 | 11.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.9 | 28.9 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 1.9 | 7.4 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 1.7 | 12.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 1.7 | 5.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 1.7 | 5.0 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 1.7 | 8.3 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 1.7 | 9.9 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 1.6 | 4.9 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 1.6 | 3.2 | GO:0006113 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 1.6 | 4.8 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 1.6 | 9.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 1.6 | 21.9 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 1.5 | 10.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 1.4 | 4.3 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 1.4 | 4.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 1.4 | 4.2 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 1.4 | 5.5 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 1.3 | 9.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 1.3 | 5.0 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 1.2 | 16.2 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 1.2 | 3.7 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 1.2 | 6.0 | GO:0061324 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 1.2 | 1.2 | GO:0072053 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 1.1 | 6.8 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 1.1 | 3.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 1.1 | 3.3 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 1.0 | 3.1 | GO:2000053 | trachea cartilage morphogenesis(GO:0060535) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 1.0 | 3.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 1.0 | 12.9 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 1.0 | 8.9 | GO:0046959 | habituation(GO:0046959) |
| 1.0 | 3.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.9 | 6.5 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.9 | 8.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.9 | 2.7 | GO:0021593 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.9 | 2.7 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.9 | 12.5 | GO:0042635 | positive regulation of hair cycle(GO:0042635) |
| 0.9 | 4.4 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.9 | 5.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.8 | 6.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.8 | 7.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.8 | 2.4 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.8 | 2.3 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.8 | 2.3 | GO:0071072 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) phenotypic switching(GO:0036166) negative regulation of phospholipid biosynthetic process(GO:0071072) metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of phenotypic switching(GO:1900239) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.8 | 2.3 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.7 | 1.5 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.7 | 6.6 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.7 | 3.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.7 | 2.8 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.7 | 2.8 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.7 | 2.8 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.7 | 3.5 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.7 | 11.0 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.7 | 6.8 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.7 | 2.7 | GO:0002159 | desmosome assembly(GO:0002159) maintenance of organ identity(GO:0048496) |
| 0.7 | 2.7 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.7 | 9.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) inner dynein arm assembly(GO:0036159) |
| 0.7 | 6.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.7 | 5.3 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.7 | 4.6 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.7 | 4.0 | GO:0061205 | alveolar primary septum development(GO:0061143) paramesonephric duct development(GO:0061205) |
| 0.6 | 4.5 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.6 | 8.9 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.6 | 1.9 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.6 | 19.6 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.6 | 3.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.6 | 2.5 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.6 | 9.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.6 | 3.1 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.6 | 2.4 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.6 | 6.6 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.6 | 2.4 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.6 | 2.8 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.5 | 19.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.5 | 3.3 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.5 | 6.0 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.5 | 2.7 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.5 | 2.1 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 0.5 | 2.6 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.5 | 2.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.5 | 2.0 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.5 | 6.1 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.5 | 3.5 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.5 | 2.0 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.5 | 6.4 | GO:0035878 | nail development(GO:0035878) |
| 0.5 | 13.7 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.5 | 0.5 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.5 | 1.4 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.5 | 6.3 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.5 | 4.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.5 | 6.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.5 | 8.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.4 | 4.0 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.4 | 12.0 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.4 | 4.9 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.4 | 7.0 | GO:0048535 | lymph node development(GO:0048535) |
| 0.4 | 2.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.4 | 1.3 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.4 | 3.8 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.4 | 2.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.4 | 2.5 | GO:0060482 | lobar bronchus epithelium development(GO:0060481) lobar bronchus development(GO:0060482) |
| 0.4 | 2.5 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.4 | 5.3 | GO:0060842 | blood vessel endothelial cell differentiation(GO:0060837) arterial endothelial cell differentiation(GO:0060842) |
| 0.4 | 0.8 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.4 | 6.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 3.4 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.4 | 4.9 | GO:0000050 | urea cycle(GO:0000050) |
| 0.4 | 8.1 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.4 | 3.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.4 | 2.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.4 | 1.5 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.3 | 2.1 | GO:1990034 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.3 | 3.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 1.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 2.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 2.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 13.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 2.5 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.3 | 4.7 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 | 6.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 3.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.3 | 9.2 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.3 | 4.5 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.3 | 8.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.3 | 0.8 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.3 | 6.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 5.8 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.3 | 2.7 | GO:0006591 | ornithine metabolic process(GO:0006591) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.3 | 1.9 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.3 | 1.3 | GO:0032909 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.3 | 4.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.3 | 1.3 | GO:0070092 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.3 | 1.6 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.3 | 3.4 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.2 | 4.1 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.2 | 11.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.2 | 2.4 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.2 | 2.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 4.0 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 5.7 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.2 | 2.5 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.2 | 1.8 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.2 | 2.2 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 | 3.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 | 4.4 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.2 | 4.6 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.2 | 5.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.2 | 7.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 0.6 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 | 0.6 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 14.7 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.2 | 4.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 3.1 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.2 | 2.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.2 | 3.2 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.2 | 1.4 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.2 | 0.6 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 2.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 3.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.2 | 3.1 | GO:0050881 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.2 | 16.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.2 | 4.2 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.2 | 3.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 4.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 0.6 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 | 8.6 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.2 | 1.9 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.2 | 3.2 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.2 | 8.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 3.6 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.2 | 5.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.2 | 0.3 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.2 | 3.6 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.2 | 2.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.2 | 3.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 22.6 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.2 | 1.5 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 9.7 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.2 | 1.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 1.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.2 | 25.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.2 | 5.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.2 | 3.7 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.2 | 3.7 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.2 | 0.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 4.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 3.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 0.3 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.2 | 2.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 13.1 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.1 | 1.9 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 16.5 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.3 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 2.5 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 1.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 3.7 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 1.9 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 2.2 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.1 | 6.4 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.1 | 0.4 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 4.2 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 5.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 7.9 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.1 | 1.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 2.9 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.1 | 2.4 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 5.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 6.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 0.9 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 3.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 2.4 | GO:2000310 | cellular response to hydroperoxide(GO:0071447) regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 4.4 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.1 | 1.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 4.6 | GO:0098773 | hair follicle development(GO:0001942) skin epidermis development(GO:0098773) |
| 0.1 | 5.4 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 2.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 4.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 3.2 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) |
| 0.1 | 1.6 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.1 | 16.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 0.8 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.1 | 2.9 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 5.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.0 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 1.2 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 15.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 6.4 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 3.3 | GO:0009798 | axis specification(GO:0009798) |
| 0.1 | 0.7 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 3.5 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 4.0 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 2.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 4.3 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 3.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 1.4 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.1 | 7.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.4 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.1 | 1.2 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.1 | 2.3 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 2.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.2 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.1 | 3.8 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 1.6 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.1 | 1.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 7.0 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 2.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 4.8 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 2.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 1.9 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 2.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.3 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 5.0 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 3.4 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 1.0 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 1.9 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.0 | 0.9 | GO:0002718 | regulation of cytokine production involved in immune response(GO:0002718) |
| 0.0 | 0.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 1.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 4.4 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 1.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 3.6 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 2.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 2.5 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.8 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.5 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 5.4 | GO:0050900 | leukocyte migration(GO:0050900) |
| 0.0 | 0.1 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 3.1 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.8 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.3 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 1.3 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.6 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 87.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 3.6 | 49.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 3.4 | 27.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 3.2 | 66.4 | GO:0005861 | troponin complex(GO:0005861) |
| 2.4 | 28.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 2.0 | 10.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 1.7 | 5.0 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 1.6 | 14.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.5 | 58.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.4 | 8.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 1.4 | 12.2 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 1.3 | 3.8 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 1.2 | 9.4 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 1.1 | 10.3 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 1.0 | 6.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 1.0 | 2.9 | GO:0043260 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.9 | 11.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.9 | 5.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.8 | 3.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.8 | 3.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.8 | 3.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.7 | 14.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.6 | 1.9 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.6 | 7.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.6 | 6.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.6 | 6.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.6 | 6.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.5 | 21.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.5 | 4.8 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.5 | 15.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.5 | 61.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.5 | 3.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.5 | 1.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.5 | 11.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.4 | 14.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 3.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.4 | 9.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.4 | 3.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.4 | 6.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.4 | 12.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.4 | 2.0 | GO:0097169 | NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.4 | 2.0 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.4 | 11.8 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.4 | 12.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.4 | 8.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.4 | 2.9 | GO:0000243 | commitment complex(GO:0000243) |
| 0.3 | 2.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.3 | 13.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 5.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 2.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.3 | 4.6 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.3 | 4.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 2.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.3 | 1.9 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.3 | 9.1 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.3 | 3.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.3 | 3.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 11.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 13.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 7.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 4.1 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 4.0 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 5.0 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.2 | 4.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.2 | 2.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 1.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 4.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 4.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 20.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.2 | 0.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 5.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 1.9 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 3.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 1.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 5.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 10.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.3 | GO:0044292 | dendrite terminus(GO:0044292) dendritic growth cone(GO:0044294) |
| 0.1 | 0.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.6 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 2.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 18.9 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 16.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 4.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 10.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 1.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 2.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 10.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 1.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 5.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 8.1 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 4.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 2.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 8.9 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.0 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.0 | 13.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 6.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.3 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 2.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 8.4 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 5.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 20.2 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 5.6 | 16.9 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 4.2 | 38.2 | GO:0043426 | MRF binding(GO:0043426) |
| 3.4 | 48.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 3.4 | 13.6 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 3.3 | 19.7 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 2.4 | 7.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 2.3 | 18.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 2.1 | 8.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 2.0 | 9.9 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 1.8 | 12.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.7 | 5.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.7 | 6.8 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 1.7 | 18.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 1.6 | 8.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 1.5 | 6.0 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 1.5 | 20.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 1.5 | 7.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.5 | 17.6 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 1.5 | 10.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 1.4 | 20.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.4 | 4.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 1.4 | 4.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 1.4 | 4.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.3 | 5.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 1.3 | 20.5 | GO:0019841 | retinol binding(GO:0019841) |
| 1.3 | 8.8 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 1.2 | 9.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 1.1 | 6.7 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 1.1 | 9.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 1.0 | 7.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 1.0 | 7.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 1.0 | 105.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 1.0 | 3.1 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 1.0 | 6.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.0 | 12.9 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.9 | 2.8 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.9 | 9.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.9 | 5.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.9 | 4.4 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.9 | 2.6 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.9 | 12.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.9 | 7.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.8 | 4.0 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.8 | 9.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.7 | 10.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.7 | 14.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.7 | 11.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.7 | 42.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.7 | 2.1 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.7 | 3.4 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.7 | 2.0 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.7 | 4.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.7 | 3.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.6 | 10.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.6 | 2.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.6 | 3.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.6 | 12.1 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.5 | 2.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.5 | 2.7 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.5 | 15.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.5 | 1.6 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.5 | 3.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.5 | 2.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.5 | 2.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 6.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 6.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 2.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.5 | 8.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.5 | 1.9 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.5 | 12.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.5 | 4.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.4 | 3.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.4 | 14.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 2.7 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 4.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 8.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 5.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 4.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 5.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.4 | 11.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.4 | 2.5 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.4 | 1.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 37.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 2.8 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.3 | 2.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.3 | 5.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.3 | 10.2 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.3 | 6.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 8.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 5.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 6.4 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.3 | 5.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.3 | 2.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.3 | 2.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 6.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 11.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.3 | 11.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.3 | 15.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.3 | 2.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 11.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 8.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.3 | 5.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.3 | 3.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 3.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 2.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 3.1 | GO:0016594 | glycine binding(GO:0016594) |
| 0.2 | 1.0 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 3.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 28.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 2.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 2.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 4.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 0.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 1.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 2.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 7.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 2.0 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 18.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 4.5 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.2 | 2.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 12.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 2.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 3.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 3.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 3.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 2.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 0.9 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 3.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 2.0 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 1.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 7.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 8.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 4.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.2 | 1.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 39.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.2 | 4.0 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.2 | 2.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 3.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 8.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 2.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 3.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 17.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 7.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 25.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 3.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.7 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 2.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 3.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 9.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.0 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 2.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 2.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 7.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 3.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.3 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 3.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 4.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 4.0 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 3.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 10.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 4.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.7 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 1.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 1.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 22.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 2.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 4.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 4.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 6.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 2.1 | GO:0001205 | RNA polymerase II repressing transcription factor binding(GO:0001103) transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 15.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 33.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 1.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.9 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.4 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 1.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 3.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.9 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 1.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 3.1 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 2.7 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 12.2 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.6 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 8.4 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) single-stranded DNA-dependent ATPase activity(GO:0043142) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 20.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.9 | 40.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.4 | 7.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.4 | 11.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 3.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 9.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.3 | 5.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 9.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.3 | 7.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 7.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.3 | 4.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 11.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.3 | 7.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 11.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 7.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 3.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 12.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.2 | 4.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 9.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 33.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 50.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 4.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 9.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 6.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 4.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 3.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 24.3 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.2 | 9.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 7.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.2 | 5.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 4.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 12.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 3.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 3.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 2.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 5.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 4.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 3.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 4.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 1.9 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 6.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.4 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 1.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 3.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 18.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 7.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 0.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 6.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 4.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 2.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 159.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.6 | 37.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.2 | 2.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 1.0 | 28.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.8 | 26.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.7 | 15.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.6 | 19.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.4 | 9.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.4 | 5.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 7.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.4 | 6.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 16.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.4 | 4.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.4 | 12.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 8.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 14.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 22.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 8.6 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.3 | 16.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 10.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 11.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 16.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.3 | 10.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 8.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.3 | 2.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.2 | 2.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 6.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 4.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 3.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 4.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 3.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 6.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.2 | 10.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.2 | 3.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 14.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 3.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 1.8 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.2 | 1.9 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 2.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 7.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.2 | 3.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 14.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 5.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 12.8 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.1 | 2.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 4.0 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 6.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 4.7 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 2.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 9.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 3.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.7 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 3.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 10.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 4.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 9.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 3.3 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 2.5 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.4 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |