Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
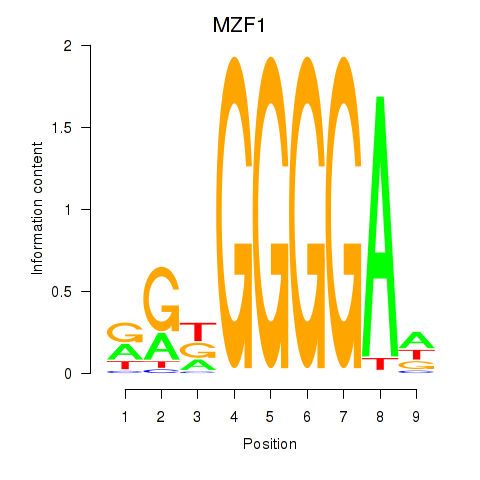
Results for MZF1
Z-value: 5.69
Transcription factors associated with MZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MZF1
|
ENSG00000099326.9 | MZF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MZF1 | hg38_v1_chr19_-_58573280_58573354, hg38_v1_chr19_-_58573555_58573582 | 0.12 | 8.8e-02 | Click! |
Activity profile of MZF1 motif
Sorted Z-values of MZF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MZF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.5 | 214.6 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 12.2 | 36.7 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 11.8 | 47.1 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 9.8 | 137.8 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 9.5 | 47.7 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 9.3 | 27.8 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 8.8 | 88.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 8.5 | 34.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 7.5 | 37.5 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 7.1 | 28.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 6.6 | 19.9 | GO:0090176 | microtubule cytoskeleton organization involved in establishment of planar polarity(GO:0090176) |
| 6.3 | 31.5 | GO:0006344 | optic cup formation involved in camera-type eye development(GO:0003408) maintenance of chromatin silencing(GO:0006344) |
| 5.3 | 10.6 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 4.9 | 24.7 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 4.5 | 26.7 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 4.0 | 12.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) regulation of chromosome condensation(GO:0060623) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 4.0 | 35.7 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 3.9 | 11.8 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 3.7 | 11.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 3.7 | 18.4 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 3.6 | 21.7 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 3.5 | 10.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 3.5 | 14.0 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 3.3 | 23.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 3.3 | 26.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 3.3 | 9.8 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 3.2 | 9.7 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 3.2 | 9.5 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 3.2 | 12.6 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 3.2 | 9.5 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 3.1 | 18.6 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 3.0 | 38.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 3.0 | 29.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 2.9 | 20.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 2.9 | 8.7 | GO:2001166 | histone H3-K79 methylation(GO:0034729) regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 2.8 | 19.9 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 2.8 | 8.5 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 2.8 | 5.6 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 2.6 | 10.4 | GO:1904799 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 2.6 | 18.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 2.6 | 10.3 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 2.5 | 17.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 2.4 | 7.3 | GO:0043465 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 2.3 | 20.5 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 2.3 | 29.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 2.2 | 29.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 2.1 | 8.5 | GO:0002317 | plasma cell differentiation(GO:0002317) response to isolation stress(GO:0035900) |
| 2.0 | 20.4 | GO:0072718 | response to cisplatin(GO:0072718) |
| 2.0 | 8.1 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 1.9 | 24.5 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 1.8 | 12.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 1.7 | 3.4 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 1.7 | 194.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 1.7 | 20.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 1.7 | 10.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 1.7 | 5.1 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 1.7 | 8.4 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 1.6 | 3.3 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 1.6 | 8.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 1.6 | 19.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 1.6 | 6.4 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 1.6 | 6.3 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 1.6 | 4.7 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 1.5 | 24.7 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 1.5 | 6.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 1.5 | 9.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 1.5 | 16.7 | GO:0007379 | segment specification(GO:0007379) |
| 1.5 | 7.6 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 1.5 | 15.0 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 1.5 | 4.5 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 1.5 | 16.5 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 1.5 | 11.7 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 1.4 | 9.7 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 1.3 | 6.7 | GO:0030242 | pexophagy(GO:0030242) |
| 1.3 | 7.8 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 1.3 | 6.4 | GO:0070295 | renal water absorption(GO:0070295) |
| 1.3 | 1.3 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 1.3 | 3.8 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 1.2 | 8.6 | GO:0072553 | terminal button organization(GO:0072553) |
| 1.2 | 1.2 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 1.2 | 4.8 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 1.2 | 34.8 | GO:0060148 | positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 1.2 | 8.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 1.2 | 7.0 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 1.2 | 18.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 1.1 | 9.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 1.1 | 40.3 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 1.1 | 9.9 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 1.1 | 5.4 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 1.0 | 17.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.0 | 3.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 1.0 | 18.0 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 1.0 | 4.0 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 1.0 | 8.9 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 1.0 | 8.9 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 1.0 | 16.6 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 1.0 | 25.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.9 | 5.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.9 | 18.4 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.9 | 13.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.9 | 5.4 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.9 | 5.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.9 | 4.3 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.8 | 7.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.8 | 6.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.8 | 8.8 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.8 | 2.4 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.8 | 2.4 | GO:0007174 | epidermal growth factor catabolic process(GO:0007174) |
| 0.8 | 9.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.8 | 5.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.8 | 2.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.8 | 5.4 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.7 | 2.2 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.7 | 19.3 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.7 | 5.7 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.7 | 0.7 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.7 | 24.4 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.7 | 2.8 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.7 | 2.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.7 | 7.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.7 | 17.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.7 | 5.9 | GO:0089700 | protein kinase D signaling(GO:0089700) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.7 | 8.5 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.7 | 3.3 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.6 | 11.7 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.6 | 1.9 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.6 | 1.9 | GO:0061445 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 0.6 | 17.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.6 | 4.5 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.6 | 7.6 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.6 | 6.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.6 | 5.5 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.6 | 2.4 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.6 | 2.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.6 | 1.2 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.6 | 9.4 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.6 | 13.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.6 | 2.9 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.6 | 4.0 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.6 | 3.9 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.6 | 13.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.5 | 6.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.5 | 1.6 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.5 | 3.7 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.5 | 7.9 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.5 | 10.2 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.5 | 8.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.5 | 0.5 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) |
| 0.5 | 6.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.5 | 1.5 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.5 | 1.5 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.5 | 5.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.5 | 1.4 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.5 | 7.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.5 | 19.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.5 | 4.6 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.5 | 2.3 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.4 | 15.7 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.4 | 4.0 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 4.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.4 | 3.0 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.4 | 2.6 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.4 | 4.8 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.4 | 11.6 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.4 | 6.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.4 | 19.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.4 | 2.0 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.4 | 5.6 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.4 | 2.0 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.4 | 0.8 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.4 | 3.9 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.4 | 28.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.4 | 3.0 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.4 | 1.5 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.4 | 3.0 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.4 | 5.1 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.4 | 2.5 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.3 | 2.4 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.3 | 3.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.3 | 1.7 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.3 | 31.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.3 | 2.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 3.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 2.3 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.3 | 6.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 8.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.3 | 0.6 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.3 | 0.9 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.3 | 0.6 | GO:0060947 | cardiac vascular smooth muscle cell differentiation(GO:0060947) |
| 0.3 | 1.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 2.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.3 | 1.8 | GO:0001781 | neutrophil apoptotic process(GO:0001781) regulation of neutrophil apoptotic process(GO:0033029) |
| 0.3 | 0.9 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.3 | 2.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 1.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.3 | 21.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.3 | 7.4 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.3 | 31.3 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.3 | 1.6 | GO:0003263 | cardioblast proliferation(GO:0003263) regulation of cardioblast proliferation(GO:0003264) |
| 0.3 | 3.2 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.3 | 9.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.3 | 0.3 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.3 | 4.8 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.3 | 8.8 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.2 | 3.0 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.2 | 24.8 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.2 | 1.0 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.2 | 2.4 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.2 | 2.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.2 | 6.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.2 | 2.1 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.2 | 0.7 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.2 | 5.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.2 | 1.6 | GO:0090666 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 38.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.2 | 1.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 1.7 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 | 1.3 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 1.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 11.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 3.5 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.2 | 7.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 2.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 9.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.2 | 0.7 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.2 | 5.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 6.0 | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition(GO:0045841) mitotic spindle checkpoint(GO:0071174) |
| 0.2 | 0.7 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 1.6 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 1.1 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 1.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.2 | 9.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 2.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.1 | 13.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 2.5 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.1 | 3.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 6.4 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 2.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 1.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 3.5 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 0.8 | GO:0060318 | regulation of definitive erythrocyte differentiation(GO:0010724) definitive erythrocyte differentiation(GO:0060318) |
| 0.1 | 4.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 2.1 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 2.9 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.1 | 0.3 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 1.8 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 3.9 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.1 | 1.9 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.7 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 3.2 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 2.4 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 6.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 8.3 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 1.5 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.1 | 4.3 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 3.9 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 0.8 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.7 | GO:0042559 | folic acid-containing compound biosynthetic process(GO:0009396) pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.1 | 4.1 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 2.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 6.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 2.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 4.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 1.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 2.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 6.1 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.9 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 6.2 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.8 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.0 | 0.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.3 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 2.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.0 | 3.8 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 1.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.1 | GO:2000739 | regulation of mesenchymal stem cell differentiation(GO:2000739) positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0060129 | corticotropin hormone secreting cell differentiation(GO:0060128) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 2.3 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 4.7 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.0 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.6 | 233.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 13.5 | 121.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 7.9 | 23.7 | GO:0008623 | CHRAC(GO:0008623) |
| 7.3 | 65.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 5.5 | 60.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 4.9 | 19.8 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 4.7 | 28.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 4.6 | 18.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 4.3 | 82.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 4.3 | 17.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 3.8 | 26.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 2.8 | 8.5 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 2.8 | 22.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 2.8 | 19.9 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 2.4 | 9.7 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 2.4 | 7.2 | GO:0072487 | MSL complex(GO:0072487) |
| 2.4 | 7.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 2.2 | 33.5 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 2.2 | 6.7 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 2.1 | 10.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 2.0 | 12.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 1.8 | 9.2 | GO:0033503 | HULC complex(GO:0033503) |
| 1.7 | 32.8 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 1.6 | 3.3 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 1.6 | 9.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 1.6 | 9.8 | GO:0097422 | tubular endosome(GO:0097422) |
| 1.5 | 10.6 | GO:0031415 | NatA complex(GO:0031415) |
| 1.5 | 7.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 1.5 | 37.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 1.5 | 16.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 1.5 | 17.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 1.5 | 16.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 1.4 | 12.7 | GO:0001739 | sex chromatin(GO:0001739) |
| 1.4 | 22.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 1.3 | 37.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 1.3 | 12.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 1.3 | 4.0 | GO:0032116 | SMC loading complex(GO:0032116) |
| 1.3 | 5.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 1.3 | 13.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 1.2 | 13.5 | GO:0032039 | integrator complex(GO:0032039) |
| 1.2 | 39.6 | GO:0071564 | SWI/SNF complex(GO:0016514) npBAF complex(GO:0071564) |
| 1.2 | 12.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 1.1 | 5.7 | GO:0036398 | TCR signalosome(GO:0036398) |
| 1.1 | 17.3 | GO:0034709 | methylosome(GO:0034709) |
| 1.1 | 13.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 1.0 | 9.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.0 | 28.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 1.0 | 8.0 | GO:0035976 | AP1 complex(GO:0035976) |
| 1.0 | 11.7 | GO:0090543 | Flemming body(GO:0090543) |
| 0.9 | 18.0 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.9 | 11.9 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.9 | 6.0 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.8 | 23.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.8 | 24.7 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.8 | 8.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.8 | 2.4 | GO:0032419 | extrinsic component of lysosome membrane(GO:0032419) |
| 0.8 | 5.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.8 | 2.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.7 | 6.6 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.7 | 10.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.7 | 12.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.7 | 7.9 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.6 | 5.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.6 | 33.1 | GO:0005844 | polysome(GO:0005844) |
| 0.6 | 7.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.6 | 25.9 | GO:0002102 | podosome(GO:0002102) |
| 0.6 | 9.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.6 | 121.1 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.6 | 7.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.5 | 21.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.5 | 3.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.5 | 22.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.5 | 1.9 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.5 | 6.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.5 | 4.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.4 | 4.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.4 | 10.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.4 | 60.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.4 | 10.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.4 | 42.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.4 | 20.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.4 | 18.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.4 | 19.4 | GO:1902493 | protein acetyltransferase complex(GO:0031248) acetyltransferase complex(GO:1902493) |
| 0.4 | 1.4 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.3 | 19.1 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.3 | 66.3 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.3 | 0.7 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 1.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 30.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.3 | 25.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 4.4 | GO:0032059 | bleb(GO:0032059) |
| 0.3 | 2.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 9.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.3 | 7.1 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.3 | 2.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.3 | 1.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.3 | 3.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 6.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.2 | 2.6 | GO:0042587 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 9.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 8.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 4.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 2.7 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 8.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 15.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 46.2 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.2 | 3.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 15.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 19.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 11.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 8.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 4.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 2.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 4.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 1.6 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.2 | 3.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 2.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.7 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 2.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 5.4 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 1.7 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 11.1 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 1.5 | GO:0033643 | host cell part(GO:0033643) |
| 0.1 | 3.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 5.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 10.7 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 9.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 9.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 11.3 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 4.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 1.8 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 9.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 7.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 2.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 4.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 3.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 6.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.6 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 10.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.0 | 1.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.5 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 8.2 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.3 | 214.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 8.5 | 34.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 7.3 | 7.3 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 6.7 | 47.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 6.2 | 18.6 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 4.3 | 25.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 4.1 | 52.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 4.0 | 11.9 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 3.9 | 11.6 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 3.7 | 143.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 3.5 | 24.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 3.3 | 26.7 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 3.3 | 26.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 3.2 | 22.5 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 3.2 | 9.6 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 3.2 | 28.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 3.1 | 28.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 3.1 | 18.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 2.9 | 23.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 2.9 | 17.3 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 2.8 | 8.5 | GO:0048030 | disaccharide binding(GO:0048030) |
| 2.5 | 17.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 2.4 | 16.9 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 2.4 | 98.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 2.3 | 16.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 2.2 | 6.6 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 2.0 | 10.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 2.0 | 13.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 1.9 | 5.6 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 1.8 | 105.4 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 1.8 | 25.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 1.8 | 23.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 1.8 | 42.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 1.7 | 8.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 1.6 | 17.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.6 | 4.8 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 1.6 | 12.6 | GO:0050733 | RS domain binding(GO:0050733) |
| 1.6 | 48.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 1.6 | 9.4 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.6 | 4.7 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 1.6 | 55.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 1.5 | 10.6 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 1.5 | 28.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 1.4 | 5.7 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 1.4 | 19.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.3 | 4.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 1.3 | 8.9 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 1.3 | 11.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 1.2 | 31.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 1.2 | 20.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 1.2 | 7.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 1.2 | 22.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 1.2 | 4.8 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 1.2 | 3.5 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 1.2 | 19.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 1.2 | 9.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.1 | 4.5 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 1.1 | 4.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.1 | 5.6 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 1.1 | 5.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 1.1 | 10.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 1.1 | 50.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 1.1 | 52.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 1.0 | 18.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 1.0 | 3.0 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 1.0 | 3.9 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 1.0 | 3.9 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.9 | 18.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.9 | 17.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.9 | 33.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.8 | 9.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.8 | 6.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.8 | 4.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.8 | 4.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.8 | 8.9 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.8 | 4.0 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.8 | 4.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.8 | 7.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.8 | 10.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.7 | 16.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.7 | 8.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.7 | 2.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.7 | 8.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.7 | 24.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.7 | 24.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.7 | 13.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.7 | 3.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.6 | 11.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.6 | 34.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.6 | 12.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.6 | 18.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.6 | 8.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.6 | 2.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.6 | 1.7 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.6 | 11.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.5 | 6.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.5 | 5.4 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 23.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.5 | 2.6 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.5 | 8.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 4.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.5 | 3.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.5 | 3.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.5 | 21.6 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.5 | 4.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.5 | 22.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.5 | 68.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.5 | 9.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.5 | 3.3 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.5 | 10.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 1.9 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.5 | 24.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.4 | 14.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.4 | 10.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 6.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.4 | 8.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 2.0 | GO:0097617 | annealing activity(GO:0097617) |
| 0.4 | 3.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.4 | 6.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.4 | 7.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.4 | 2.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 1.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 69.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.3 | 2.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 8.1 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 13.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 11.2 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.3 | 1.9 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 19.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.3 | 1.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 7.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.3 | 4.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 19.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.3 | 1.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.3 | 10.0 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.3 | 3.0 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.3 | 16.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.3 | 1.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 6.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 5.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 16.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 0.3 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.3 | 1.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.3 | 16.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 1.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 4.5 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.2 | 1.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 2.0 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 1.5 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 19.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.2 | 2.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 5.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 0.7 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.2 | 2.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 9.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 4.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 7.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 0.8 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.2 | 18.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.2 | 15.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 1.6 | GO:0046923 | A-type (transient outward) potassium channel activity(GO:0005250) ER retention sequence binding(GO:0046923) |
| 0.2 | 10.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 2.6 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.2 | 8.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.2 | 3.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 2.7 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 5.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.2 | 28.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 3.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 5.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 1.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 5.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 7.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 2.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.7 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 1.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 1.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 6.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.3 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 9.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 3.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.8 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 2.0 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 4.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 16.1 | GO:0061659 | ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 1.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 6.1 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 3.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 10.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 2.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.7 | GO:0031701 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.8 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 1.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 3.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.0 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 214.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 3.0 | 56.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 2.5 | 86.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 1.3 | 26.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 1.2 | 43.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 1.2 | 32.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.9 | 46.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.9 | 20.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.8 | 48.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.8 | 5.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.7 | 51.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.7 | 75.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.7 | 28.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.7 | 20.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.6 | 50.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.5 | 47.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.5 | 33.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.5 | 41.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.5 | 29.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.4 | 25.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.4 | 18.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.4 | 21.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.3 | 10.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.3 | 14.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.3 | 30.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.3 | 21.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.3 | 3.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 17.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 4.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 17.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 5.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 3.2 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 3.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.2 | 3.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 6.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 5.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 6.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 4.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 4.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 2.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 4.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 4.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 11.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 2.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 1.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 218.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 3.1 | 47.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 2.5 | 50.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 1.6 | 1.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 1.5 | 36.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 1.4 | 23.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 1.3 | 14.6 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 1.2 | 25.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 1.1 | 8.9 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 1.1 | 9.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 1.1 | 8.5 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 1.0 | 20.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 1.0 | 31.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.9 | 17.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.9 | 46.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.9 | 93.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.8 | 11.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.8 | 18.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.8 | 39.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.8 | 26.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.7 | 22.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.7 | 13.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.6 | 22.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.6 | 23.9 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.6 | 9.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.6 | 20.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.5 | 11.9 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.5 | 5.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.5 | 36.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.5 | 83.2 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.5 | 12.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.5 | 12.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.5 | 18.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.4 | 20.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.4 | 6.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.4 | 5.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.4 | 7.8 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.3 | 15.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.3 | 8.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 12.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.3 | 4.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.3 | 9.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 17.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 1.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.3 | 3.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 6.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 6.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 6.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 4.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.2 | 4.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 6.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 6.5 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.2 | 5.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 7.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.2 | 14.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 32.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.2 | 6.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 8.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 6.6 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.2 | 12.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 12.4 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.2 | 2.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 3.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 5.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 5.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 6.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 4.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.4 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 4.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 8.5 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 2.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.1 | 13.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 6.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 1.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 0.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.6 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |