Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for NANOG
Z-value: 0.57
Transcription factors associated with NANOG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NANOG
|
ENSG00000111704.11 | NANOG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NANOG | hg38_v1_chr12_+_7789393_7789417 | -0.02 | 7.5e-01 | Click! |
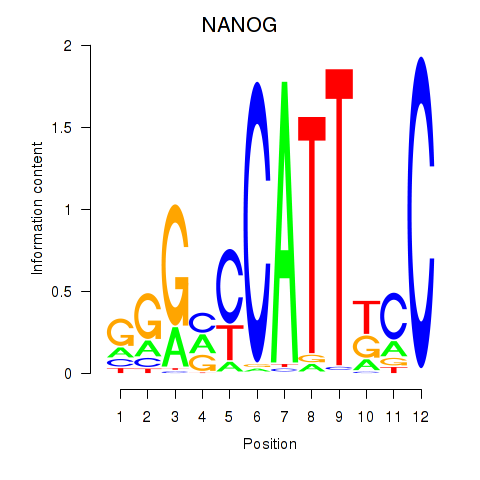
Activity profile of NANOG motif
Sorted Z-values of NANOG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NANOG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_38852321 | 4.79 |
ENST00000600873.5
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L |
| chr5_+_157266079 | 4.11 |
ENST00000616178.4
ENST00000522463.5 ENST00000435847.6 ENST00000620254.5 ENST00000521420.5 ENST00000617629.4 |
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr11_-_67437670 | 3.33 |
ENST00000326294.4
|
PTPRCAP
|
protein tyrosine phosphatase receptor type C associated protein |
| chrX_-_119693150 | 3.12 |
ENST00000394610.7
|
SEPTIN6
|
septin 6 |
| chr10_+_10798570 | 3.09 |
ENST00000638035.1
ENST00000636488.1 |
CELF2
|
CUGBP Elav-like family member 2 |
| chr17_-_36090133 | 2.59 |
ENST00000613922.2
|
CCL3
|
C-C motif chemokine ligand 3 |
| chrX_-_119693370 | 2.58 |
ENST00000360156.11
ENST00000354228.8 ENST00000489216.5 ENST00000354416.7 ENST00000343984.5 |
SEPTIN6
|
septin 6 |
| chr12_-_121793668 | 2.43 |
ENST00000267205.7
|
RHOF
|
ras homolog family member F, filopodia associated |
| chr12_-_122703346 | 2.42 |
ENST00000328880.6
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr19_-_42302576 | 2.31 |
ENST00000262890.8
|
PAFAH1B3
|
platelet activating factor acetylhydrolase 1b catalytic subunit 3 |
| chr19_-_42302766 | 2.27 |
ENST00000595530.5
ENST00000538771.5 ENST00000601865.5 |
PAFAH1B3
|
platelet activating factor acetylhydrolase 1b catalytic subunit 3 |
| chr10_+_128047559 | 2.16 |
ENST00000306042.9
|
PTPRE
|
protein tyrosine phosphatase receptor type E |
| chr19_-_42302292 | 2.14 |
ENST00000594989.5
|
PAFAH1B3
|
platelet activating factor acetylhydrolase 1b catalytic subunit 3 |
| chr19_-_42302690 | 2.13 |
ENST00000596265.5
|
PAFAH1B3
|
platelet activating factor acetylhydrolase 1b catalytic subunit 3 |
| chr11_+_126304041 | 2.10 |
ENST00000263579.5
|
DCPS
|
decapping enzyme, scavenger |
| chr4_+_165327659 | 2.05 |
ENST00000507013.5
ENST00000261507.11 ENST00000393766.6 ENST00000504317.1 |
MSMO1
|
methylsterol monooxygenase 1 |
| chr11_-_72793592 | 1.99 |
ENST00000536377.5
ENST00000359373.9 |
STARD10
ARAP1
|
StAR related lipid transfer domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr12_-_122716790 | 1.86 |
ENST00000528880.3
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr19_+_926001 | 1.80 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr19_-_13102848 | 1.78 |
ENST00000264824.5
|
LYL1
|
LYL1 basic helix-loop-helix family member |
| chr1_+_44405247 | 1.71 |
ENST00000361799.7
ENST00000372247.6 |
RNF220
|
ring finger protein 220 |
| chr14_+_23322019 | 1.60 |
ENST00000557702.5
|
PABPN1
|
poly(A) binding protein nuclear 1 |
| chr15_+_78540729 | 1.55 |
ENST00000559365.5
ENST00000558341.5 ENST00000559437.5 |
PSMA4
|
proteasome 20S subunit alpha 4 |
| chr8_-_90082871 | 1.54 |
ENST00000265431.7
|
CALB1
|
calbindin 1 |
| chr1_+_44405164 | 1.53 |
ENST00000355387.6
|
RNF220
|
ring finger protein 220 |
| chr15_+_78540405 | 1.50 |
ENST00000560217.5
ENST00000559082.5 ENST00000559948.5 ENST00000413382.6 ENST00000559146.5 ENST00000044462.12 ENST00000558281.5 |
PSMA4
|
proteasome 20S subunit alpha 4 |
| chr14_+_23321543 | 1.50 |
ENST00000556821.5
|
PABPN1
|
poly(A) binding protein nuclear 1 |
| chr6_+_34236865 | 1.40 |
ENST00000674029.1
ENST00000447654.5 ENST00000347617.10 ENST00000401473.7 ENST00000311487.9 |
HMGA1
|
high mobility group AT-hook 1 |
| chr1_+_159302321 | 1.36 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE receptor Ia |
| chr6_-_32192630 | 1.32 |
ENST00000375040.8
|
GPSM3
|
G protein signaling modulator 3 |
| chr1_-_28193873 | 1.30 |
ENST00000305392.3
ENST00000539896.1 |
PTAFR
|
platelet activating factor receptor |
| chr9_+_69205141 | 1.26 |
ENST00000649134.1
ENST00000539225.2 ENST00000650084.1 ENST00000648402.1 |
TJP2
|
tight junction protein 2 |
| chr14_-_60165363 | 1.24 |
ENST00000557185.6
|
DHRS7
|
dehydrogenase/reductase 7 |
| chr1_+_161581339 | 1.19 |
ENST00000543859.5
ENST00000611236.1 |
FCGR2C
|
Fc fragment of IgG receptor IIc (gene/pseudogene) |
| chr16_-_11256192 | 1.19 |
ENST00000644787.1
ENST00000332029.4 |
SOCS1
|
suppressor of cytokine signaling 1 |
| chr1_+_161663147 | 1.18 |
ENST00000236937.13
ENST00000367961.8 ENST00000358671.9 |
FCGR2B
|
Fc fragment of IgG receptor IIb |
| chr19_-_39435910 | 1.11 |
ENST00000599539.5
|
RPS16
|
ribosomal protein S16 |
| chr6_-_161274010 | 1.11 |
ENST00000366911.9
ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr6_+_31665234 | 1.11 |
ENST00000677536.1
ENST00000375885.8 |
CSNK2B
|
casein kinase 2 beta |
| chr6_+_31572279 | 1.09 |
ENST00000418386.3
|
LTA
|
lymphotoxin alpha |
| chr1_-_202159977 | 1.05 |
ENST00000367279.8
|
PTPN7
|
protein tyrosine phosphatase non-receptor type 7 |
| chr6_-_161274042 | 1.04 |
ENST00000320285.9
|
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr19_-_39435627 | 1.00 |
ENST00000599705.1
|
RPS16
|
ribosomal protein S16 |
| chr6_+_30717433 | 0.98 |
ENST00000681435.1
|
TUBB
|
tubulin beta class I |
| chr22_-_41688859 | 0.97 |
ENST00000401959.6
ENST00000648674.1 |
SNU13
|
small nuclear ribonucleoprotein 13 |
| chr19_-_39435936 | 0.97 |
ENST00000339471.8
ENST00000601655.5 ENST00000251453.8 |
RPS16
|
ribosomal protein S16 |
| chr7_-_78771265 | 0.95 |
ENST00000630991.2
ENST00000629359.2 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr6_+_31571957 | 0.93 |
ENST00000454783.5
|
LTA
|
lymphotoxin alpha |
| chr10_-_27154226 | 0.92 |
ENST00000427324.5
ENST00000375972.7 ENST00000326799.7 |
YME1L1
|
YME1 like 1 ATPase |
| chr2_+_169733811 | 0.92 |
ENST00000392647.7
|
KLHL23
|
kelch like family member 23 |
| chr4_+_39698109 | 0.89 |
ENST00000510934.5
ENST00000261427.10 |
UBE2K
|
ubiquitin conjugating enzyme E2 K |
| chr22_+_37051731 | 0.86 |
ENST00000610767.4
ENST00000402077.7 ENST00000403888.7 |
KCTD17
|
potassium channel tetramerization domain containing 17 |
| chr22_+_37051787 | 0.86 |
ENST00000456470.1
|
KCTD17
|
potassium channel tetramerization domain containing 17 |
| chr6_+_30557287 | 0.85 |
ENST00000376560.8
|
PRR3
|
proline rich 3 |
| chr5_+_42548043 | 0.81 |
ENST00000618088.4
ENST00000612382.4 |
GHR
|
growth hormone receptor |
| chr10_+_96130027 | 0.81 |
ENST00000478086.5
ENST00000624776.3 |
ZNF518A
|
zinc finger protein 518A |
| chr2_-_213151590 | 0.80 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr6_+_30557274 | 0.78 |
ENST00000376557.3
|
PRR3
|
proline rich 3 |
| chr19_-_49867542 | 0.77 |
ENST00000600910.5
ENST00000322344.8 ENST00000600573.5 ENST00000596726.3 ENST00000638016.1 |
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr10_+_96129707 | 0.75 |
ENST00000316045.9
|
ZNF518A
|
zinc finger protein 518A |
| chr11_+_26332117 | 0.74 |
ENST00000531646.1
ENST00000256737.8 |
ANO3
|
anoctamin 3 |
| chr15_+_77420880 | 0.74 |
ENST00000336216.9
ENST00000558176.1 |
HMG20A
|
high mobility group 20A |
| chr3_+_122055355 | 0.73 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr2_-_174248570 | 0.73 |
ENST00000344357.9
|
OLA1
|
Obg like ATPase 1 |
| chr17_+_77376083 | 0.72 |
ENST00000427674.6
|
SEPTIN9
|
septin 9 |
| chr17_-_45051517 | 0.72 |
ENST00000614054.4
|
DCAKD
|
dephospho-CoA kinase domain containing |
| chr17_-_45051575 | 0.72 |
ENST00000588499.5
ENST00000593094.1 ENST00000651974.1 |
DCAKD
|
dephospho-CoA kinase domain containing |
| chr15_+_41256907 | 0.71 |
ENST00000560965.1
|
CHP1
|
calcineurin like EF-hand protein 1 |
| chr19_+_49581304 | 0.71 |
ENST00000246794.10
|
PRRG2
|
proline rich and Gla domain 2 |
| chr1_+_62436297 | 0.70 |
ENST00000452143.5
ENST00000442679.5 ENST00000371146.5 |
USP1
|
ubiquitin specific peptidase 1 |
| chr22_-_41688799 | 0.69 |
ENST00000469028.2
ENST00000463675.6 ENST00000649479.1 ENST00000469522.1 |
SNU13
|
small nuclear ribonucleoprotein 13 |
| chr16_+_30748241 | 0.68 |
ENST00000565924.5
ENST00000424889.7 |
PHKG2
|
phosphorylase kinase catalytic subunit gamma 2 |
| chr10_+_67884646 | 0.68 |
ENST00000212015.11
|
SIRT1
|
sirtuin 1 |
| chr5_-_116536458 | 0.64 |
ENST00000510263.5
|
SEMA6A
|
semaphorin 6A |
| chr18_+_13218769 | 0.64 |
ENST00000677055.1
ENST00000399848.7 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr7_-_78771108 | 0.62 |
ENST00000626691.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr16_+_30748378 | 0.61 |
ENST00000563588.6
ENST00000328273.11 |
PHKG2
|
phosphorylase kinase catalytic subunit gamma 2 |
| chr4_+_39698156 | 0.61 |
ENST00000503368.5
ENST00000445950.2 |
UBE2K
|
ubiquitin conjugating enzyme E2 K |
| chr22_-_30266839 | 0.60 |
ENST00000403463.1
ENST00000215781.3 |
OSM
|
oncostatin M |
| chr20_+_44714835 | 0.60 |
ENST00000372868.6
|
CCN5
|
cellular communication network factor 5 |
| chr19_-_49867251 | 0.59 |
ENST00000631020.2
ENST00000596014.5 ENST00000636994.1 |
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr22_+_26169474 | 0.59 |
ENST00000404234.7
ENST00000529632.6 ENST00000360929.7 ENST00000629590.2 ENST00000343706.8 |
SEZ6L
|
seizure related 6 homolog like |
| chr20_+_44714853 | 0.59 |
ENST00000372865.4
|
CCN5
|
cellular communication network factor 5 |
| chr12_-_89708816 | 0.56 |
ENST00000428670.8
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr7_-_77199808 | 0.56 |
ENST00000248598.6
|
FGL2
|
fibrinogen like 2 |
| chrX_-_107775951 | 0.55 |
ENST00000315660.8
ENST00000372384.6 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family member 3 |
| chr6_+_106629560 | 0.54 |
ENST00000369046.8
|
QRSL1
|
glutaminyl-tRNA amidotransferase subunit QRSL1 |
| chr20_+_44715360 | 0.52 |
ENST00000190983.5
|
CCN5
|
cellular communication network factor 5 |
| chrX_-_48971829 | 0.51 |
ENST00000218176.4
|
KCND1
|
potassium voltage-gated channel subfamily D member 1 |
| chr11_-_57237183 | 0.51 |
ENST00000606794.1
|
APLNR
|
apelin receptor |
| chr1_-_155990062 | 0.51 |
ENST00000462460.6
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr2_-_174248360 | 0.51 |
ENST00000409546.5
ENST00000428402.6 ENST00000284719.8 |
OLA1
|
Obg like ATPase 1 |
| chr4_+_113116676 | 0.50 |
ENST00000671971.1
ENST00000672240.1 ENST00000673240.1 ENST00000673363.1 |
ANK2
|
ankyrin 2 |
| chr3_-_49813880 | 0.49 |
ENST00000333486.4
|
UBA7
|
ubiquitin like modifier activating enzyme 7 |
| chr3_+_132660305 | 0.44 |
ENST00000683741.1
ENST00000468022.5 ENST00000356232.10 ENST00000473651.5 ENST00000494238.6 |
UBA5
|
ubiquitin like modifier activating enzyme 5 |
| chr22_+_26169454 | 0.44 |
ENST00000248933.11
|
SEZ6L
|
seizure related 6 homolog like |
| chr12_-_31591129 | 0.44 |
ENST00000389082.10
|
DENND5B
|
DENN domain containing 5B |
| chr10_+_122163672 | 0.41 |
ENST00000369004.7
ENST00000260733.7 |
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr9_+_134326435 | 0.39 |
ENST00000481739.2
|
RXRA
|
retinoid X receptor alpha |
| chr2_+_45651650 | 0.37 |
ENST00000306156.8
|
PRKCE
|
protein kinase C epsilon |
| chr1_-_150268941 | 0.37 |
ENST00000369109.8
ENST00000236017.5 |
APH1A
|
aph-1 homolog A, gamma-secretase subunit |
| chr17_+_8002610 | 0.37 |
ENST00000254854.5
|
GUCY2D
|
guanylate cyclase 2D, retinal |
| chr1_+_24643264 | 0.34 |
ENST00000374389.8
ENST00000323848.14 ENST00000447431.6 |
SRRM1
|
serine and arginine repetitive matrix 1 |
| chr11_+_4704782 | 0.33 |
ENST00000380390.6
|
MMP26
|
matrix metallopeptidase 26 |
| chr1_+_35883189 | 0.30 |
ENST00000674304.1
ENST00000373204.6 ENST00000674426.1 |
AGO1
|
argonaute RISC component 1 |
| chr11_-_35360050 | 0.29 |
ENST00000644868.1
ENST00000643454.1 ENST00000646080.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr18_-_26090584 | 0.28 |
ENST00000415083.7
|
SS18
|
SS18 subunit of BAF chromatin remodeling complex |
| chrX_+_44844015 | 0.26 |
ENST00000339042.6
|
DUSP21
|
dual specificity phosphatase 21 |
| chr14_+_20688756 | 0.24 |
ENST00000397990.5
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin ribonuclease A family member 4 |
| chr6_-_136289824 | 0.23 |
ENST00000527536.5
ENST00000529826.5 ENST00000531224.6 ENST00000353331.8 ENST00000628517.2 |
BCLAF1
|
BCL2 associated transcription factor 1 |
| chr10_+_122163426 | 0.22 |
ENST00000360561.7
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr10_+_69124152 | 0.22 |
ENST00000395098.5
ENST00000263559.11 |
VPS26A
|
VPS26, retromer complex component A |
| chr8_+_28701487 | 0.22 |
ENST00000220562.9
|
EXTL3
|
exostosin like glycosyltransferase 3 |
| chr3_-_24494791 | 0.21 |
ENST00000431815.5
ENST00000356447.9 ENST00000418247.1 ENST00000416420.5 ENST00000396671.7 |
THRB
|
thyroid hormone receptor beta |
| chr6_+_87322581 | 0.20 |
ENST00000608353.5
ENST00000392863.6 ENST00000229570.9 ENST00000608525.5 ENST00000608868.2 |
SMIM8
|
small integral membrane protein 8 |
| chr5_+_141182369 | 0.20 |
ENST00000609684.3
ENST00000625044.1 ENST00000623407.1 ENST00000623884.1 |
PCDHB16
ENSG00000279068.1
|
protocadherin beta 16 novel transcript |
| chr1_+_203128279 | 0.19 |
ENST00000367235.1
ENST00000618295.1 |
ADORA1
|
adenosine A1 receptor |
| chr1_-_115841116 | 0.19 |
ENST00000320238.3
|
NHLH2
|
nescient helix-loop-helix 2 |
| chr1_-_147773341 | 0.17 |
ENST00000430508.1
ENST00000621517.1 |
GJA5
|
gap junction protein alpha 5 |
| chr1_+_201955496 | 0.17 |
ENST00000367287.5
|
TIMM17A
|
translocase of inner mitochondrial membrane 17A |
| chr11_+_68312542 | 0.17 |
ENST00000294304.12
|
LRP5
|
LDL receptor related protein 5 |
| chr3_+_44874606 | 0.16 |
ENST00000296125.9
|
TGM4
|
transglutaminase 4 |
| chr16_+_6019071 | 0.16 |
ENST00000547605.5
ENST00000553186.5 |
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr14_+_22052503 | 0.15 |
ENST00000390449.3
|
TRAV21
|
T cell receptor alpha variable 21 |
| chr21_-_33588624 | 0.14 |
ENST00000437395.5
ENST00000453626.5 ENST00000303113.10 ENST00000303071.10 ENST00000432378.5 |
DONSON
|
DNA replication fork stabilization factor DONSON |
| chr10_-_48604952 | 0.14 |
ENST00000417912.6
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr16_+_6019016 | 0.13 |
ENST00000550418.6
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr10_-_67696115 | 0.12 |
ENST00000433211.7
|
CTNNA3
|
catenin alpha 3 |
| chr12_-_51083582 | 0.12 |
ENST00000548206.1
ENST00000546935.5 ENST00000228515.6 ENST00000548981.5 |
CSRNP2
|
cysteine and serine rich nuclear protein 2 |
| chr10_+_101131284 | 0.12 |
ENST00000370196.11
ENST00000467928.2 |
TLX1
|
T cell leukemia homeobox 1 |
| chr11_+_34051722 | 0.11 |
ENST00000341394.9
ENST00000389645.7 |
CAPRIN1
|
cell cycle associated protein 1 |
| chr10_-_48605032 | 0.10 |
ENST00000249601.9
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr17_+_50095331 | 0.10 |
ENST00000503176.6
|
PDK2
|
pyruvate dehydrogenase kinase 2 |
| chr1_-_145859043 | 0.09 |
ENST00000369298.5
|
PIAS3
|
protein inhibitor of activated STAT 3 |
| chr10_+_62374361 | 0.09 |
ENST00000395254.8
|
ZNF365
|
zinc finger protein 365 |
| chr10_+_122163590 | 0.09 |
ENST00000368999.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr1_-_145859061 | 0.09 |
ENST00000393045.7
|
PIAS3
|
protein inhibitor of activated STAT 3 |
| chr5_-_95284535 | 0.09 |
ENST00000515393.5
|
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr3_-_24494875 | 0.09 |
ENST00000644321.1
|
THRB
|
thyroid hormone receptor beta |
| chr3_-_24494842 | 0.09 |
ENST00000646209.2
|
THRB
|
thyroid hormone receptor beta |
| chr20_-_21514046 | 0.08 |
ENST00000377142.5
|
NKX2-2
|
NK2 homeobox 2 |
| chr7_+_6374491 | 0.08 |
ENST00000348035.9
ENST00000356142.4 |
RAC1
|
Rac family small GTPase 1 |
| chrX_+_47223009 | 0.08 |
ENST00000518022.5
ENST00000276052.10 |
CDK16
|
cyclin dependent kinase 16 |
| chr17_+_50095285 | 0.07 |
ENST00000503614.5
|
PDK2
|
pyruvate dehydrogenase kinase 2 |
| chr2_+_170816562 | 0.07 |
ENST00000625689.2
ENST00000375272.5 |
GAD1
|
glutamate decarboxylase 1 |
| chr1_+_46798998 | 0.06 |
ENST00000640628.1
ENST00000271153.8 ENST00000371923.9 ENST00000371919.8 ENST00000614163.4 |
CYP4B1
|
cytochrome P450 family 4 subfamily B member 1 |
| chr1_-_150269051 | 0.04 |
ENST00000414276.6
ENST00000360244.8 |
APH1A
|
aph-1 homolog A, gamma-secretase subunit |
| chr5_-_170389349 | 0.04 |
ENST00000274629.9
|
KCNMB1
|
potassium calcium-activated channel subfamily M regulatory beta subunit 1 |
| chr6_-_24719146 | 0.03 |
ENST00000378119.9
|
C6orf62
|
chromosome 6 open reading frame 62 |
| chr10_+_988425 | 0.03 |
ENST00000360803.9
|
GTPBP4
|
GTP binding protein 4 |
| chr9_+_137788781 | 0.01 |
ENST00000482340.5
|
EHMT1
|
euchromatic histone lysine methyltransferase 1 |
| chr20_-_62926469 | 0.00 |
ENST00000354665.8
ENST00000370368.5 ENST00000395340.5 ENST00000395343.6 |
DIDO1
|
death inducer-obliterator 1 |
| chr1_-_155188351 | 0.00 |
ENST00000462317.5
|
MUC1
|
mucin 1, cell surface associated |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.7 | 2.1 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.7 | 2.0 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.6 | 2.5 | GO:0016068 | regulation of type I hypersensitivity(GO:0001810) type I hypersensitivity(GO:0016068) |
| 0.6 | 4.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.5 | 2.0 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.5 | 2.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.5 | 1.4 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.4 | 2.6 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.3 | 1.4 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.3 | 1.3 | GO:1903238 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.2 | 0.7 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.2 | 0.7 | GO:0032761 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.2 | 0.7 | GO:0051097 | negative regulation of prostaglandin biosynthetic process(GO:0031393) negative regulation of helicase activity(GO:0051097) negative regulation of response to alcohol(GO:1901420) |
| 0.2 | 1.3 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.2 | 0.4 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.2 | 1.5 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.2 | 0.5 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.2 | 1.5 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.2 | 1.3 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.2 | 1.6 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.2 | 2.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 1.8 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 1.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 1.2 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.1 | 0.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 1.7 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.8 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.9 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.7 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.6 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 1.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 | 3.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 3.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.6 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 5.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.2 | GO:0070256 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.1 | 0.4 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 1.7 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.2 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.4 | GO:0008050 | negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.1 | 1.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.5 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.3 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.5 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 3.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.7 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0061304 | bone marrow development(GO:0048539) retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.6 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 2.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.9 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.4 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.0 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 3.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 1.8 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 7.3 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 2.1 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 1.0 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.3 | 0.8 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.3 | 3.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 1.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 2.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 0.5 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.2 | 1.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 3.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 1.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.7 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 1.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 3.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 8.6 | GO:0035770 | ribonucleoprotein granule(GO:0035770) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.5 | GO:0043198 | dendritic shaft(GO:0043198) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 8.8 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.6 | 2.6 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.6 | 1.7 | GO:0034512 | box C/D snoRNA binding(GO:0034512) |
| 0.4 | 4.8 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.3 | 1.4 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.3 | 1.3 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.3 | 2.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.3 | 1.4 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.2 | 1.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 2.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 0.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 1.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 0.5 | GO:0050567 | amidase activity(GO:0004040) glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.4 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.1 | 0.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 3.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 2.0 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 1.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.4 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.1 | 2.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 3.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 2.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.2 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.1 | 1.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 3.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 2.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 1.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0051021 | thioesterase binding(GO:0031996) GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 3.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 3.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 4.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 2.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 3.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 0.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 3.1 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 3.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 2.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 2.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 4.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 5.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 4.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 3.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |