Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for NFATC2_NFATC3
Z-value: 4.67
Transcription factors associated with NFATC2_NFATC3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
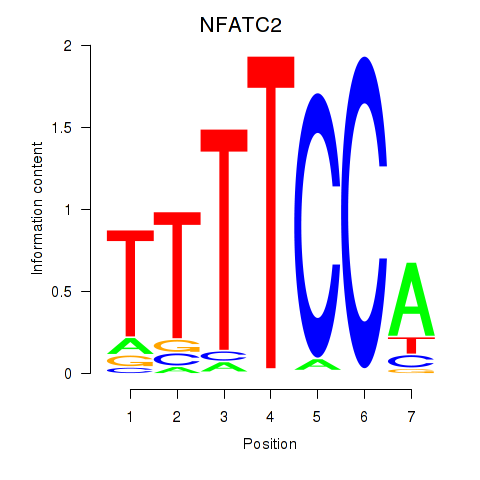
NFATC2
|
ENSG00000101096.20 | NFATC2 |
|
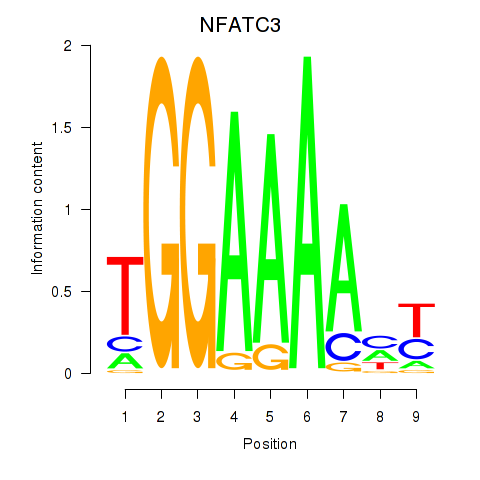
NFATC3
|
ENSG00000072736.19 | NFATC3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC3 | hg38_v1_chr16_+_68085861_68085968 | -0.64 | 1.3e-26 | Click! |
Activity profile of NFATC2_NFATC3 motif
Sorted Z-values of NFATC2_NFATC3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC2_NFATC3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.1 | 157.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 7.4 | 29.6 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 7.3 | 22.0 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 6.0 | 18.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 5.5 | 27.6 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 4.3 | 13.0 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 4.3 | 12.9 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 3.6 | 43.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 3.3 | 9.9 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 3.3 | 9.8 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 3.2 | 9.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 3.0 | 12.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 2.9 | 23.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 2.9 | 8.7 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 2.7 | 13.6 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 2.6 | 7.9 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 2.4 | 7.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 2.4 | 14.4 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 2.3 | 18.6 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 2.3 | 9.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 2.2 | 10.9 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 2.1 | 6.2 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 2.0 | 18.4 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 2.0 | 8.1 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 2.0 | 10.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 2.0 | 7.9 | GO:0009956 | radial pattern formation(GO:0009956) |
| 2.0 | 5.9 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 1.9 | 5.7 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 1.8 | 3.6 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 1.8 | 5.3 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 1.7 | 12.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.7 | 8.5 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) negative regulation of gastrulation(GO:2000542) |
| 1.7 | 6.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 1.7 | 5.0 | GO:0009386 | translational attenuation(GO:0009386) |
| 1.5 | 6.2 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 1.5 | 5.9 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 1.5 | 10.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 1.5 | 4.4 | GO:0060596 | mammary placode formation(GO:0060596) |
| 1.4 | 5.8 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 1.4 | 21.9 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 1.4 | 13.7 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.3 | 4.0 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 1.3 | 5.3 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 1.3 | 2.6 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 1.3 | 3.9 | GO:1902617 | response to fluoride(GO:1902617) |
| 1.3 | 2.6 | GO:0061027 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 1.2 | 3.7 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 1.2 | 3.7 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 1.2 | 3.7 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 1.2 | 4.8 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 1.2 | 3.6 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 1.1 | 7.8 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 1.1 | 8.9 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 1.1 | 12.3 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.1 | 3.3 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 1.0 | 3.0 | GO:1900920 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 1.0 | 15.8 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 1.0 | 2.9 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) |
| 1.0 | 3.9 | GO:0048539 | bone marrow development(GO:0048539) |
| 1.0 | 10.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.9 | 14.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.9 | 11.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.9 | 10.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.9 | 2.8 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.9 | 19.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.9 | 0.9 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.9 | 8.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.9 | 9.9 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.9 | 4.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.9 | 1.7 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.9 | 2.6 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.8 | 16.1 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.8 | 1.6 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.8 | 2.4 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.8 | 4.6 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.8 | 6.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.8 | 2.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.8 | 2.3 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.7 | 3.0 | GO:0061443 | ciliary body morphogenesis(GO:0061073) endocardial cushion cell differentiation(GO:0061443) |
| 0.7 | 1.5 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.7 | 2.9 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.7 | 2.9 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.7 | 8.6 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.7 | 5.7 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.7 | 2.1 | GO:0072313 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.7 | 2.8 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.7 | 2.8 | GO:0070091 | glucagon secretion(GO:0070091) |
| 0.7 | 2.1 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.7 | 8.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.7 | 6.1 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.7 | 4.7 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.7 | 49.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.6 | 6.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.6 | 1.9 | GO:0039008 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.6 | 16.5 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.6 | 8.0 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.6 | 1.9 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.6 | 4.9 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.6 | 7.0 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.6 | 2.3 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.6 | 2.3 | GO:1900190 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.6 | 5.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.6 | 9.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.6 | 2.9 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.6 | 2.9 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.6 | 2.8 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.6 | 6.8 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.6 | 12.4 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.5 | 6.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.5 | 4.9 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.5 | 2.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.5 | 1.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.5 | 0.5 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.5 | 3.1 | GO:0046086 | AMP catabolic process(GO:0006196) adenosine biosynthetic process(GO:0046086) |
| 0.5 | 2.6 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.5 | 3.6 | GO:2000229 | cellular response to vitamin E(GO:0071306) pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.5 | 6.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.5 | 2.5 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.5 | 0.5 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.5 | 2.5 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.5 | 5.9 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 4.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.5 | 4.4 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.5 | 1.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.5 | 1.4 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.5 | 1.8 | GO:1905229 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.5 | 1.4 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.5 | 1.8 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.5 | 13.1 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.4 | 1.8 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.4 | 1.8 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.4 | 3.5 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.4 | 4.4 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.4 | 3.7 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.4 | 5.0 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.4 | 4.9 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.4 | 64.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 | 3.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.4 | 7.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.4 | 4.0 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.4 | 8.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.4 | 3.2 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.4 | 1.2 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.4 | 2.4 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.4 | 1.9 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.4 | 3.5 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 1.9 | GO:0007497 | posterior midgut development(GO:0007497) enteric smooth muscle cell differentiation(GO:0035645) |
| 0.4 | 4.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.4 | 0.7 | GO:2000855 | renin-angiotensin regulation of aldosterone production(GO:0002018) mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.4 | 6.5 | GO:0033622 | integrin activation(GO:0033622) |
| 0.4 | 1.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.3 | 2.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.3 | 3.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.3 | 2.4 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.3 | 3.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 | 2.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 3.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.3 | 2.3 | GO:0010269 | triglyceride mobilization(GO:0006642) response to selenium ion(GO:0010269) |
| 0.3 | 2.0 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 2.6 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.3 | 7.4 | GO:0071402 | cellular response to lipoprotein particle stimulus(GO:0071402) |
| 0.3 | 10.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.3 | 3.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.3 | 4.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.3 | 5.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.3 | 0.9 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.3 | 2.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 1.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.3 | 9.9 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 | 2.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.3 | 1.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.3 | 0.8 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.3 | 7.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 1.6 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.3 | 1.6 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.3 | 0.8 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.3 | 0.8 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.3 | 1.9 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.3 | 1.6 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.3 | 1.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.3 | 0.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.3 | 4.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.3 | 14.5 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.3 | 2.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.3 | 1.5 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.2 | 3.0 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.2 | 3.2 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 1.9 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 2.6 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 2.3 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.2 | 5.9 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.2 | 3.9 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 2.5 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.2 | 1.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 3.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 15.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.2 | 26.8 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.2 | 0.8 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.2 | 5.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.2 | 1.7 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.2 | 6.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 1.8 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 2.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 | 1.4 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.2 | 1.4 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.2 | 2.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 0.6 | GO:0034241 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.2 | 2.1 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.2 | 2.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 3.4 | GO:0043586 | tongue development(GO:0043586) |
| 0.2 | 1.1 | GO:1903392 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.2 | 5.3 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.2 | 0.4 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.2 | 5.4 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.2 | 1.9 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 3.9 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.2 | 1.5 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.2 | 0.9 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.2 | 2.0 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 0.2 | 5.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 4.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 2.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 3.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.2 | 1.8 | GO:0097116 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.2 | 4.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 2.0 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 1.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 1.4 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.1 | 1.0 | GO:0046061 | dGTP catabolic process(GO:0006203) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dATP catabolic process(GO:0046061) |
| 0.1 | 0.9 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.1 | 1.0 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 3.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 2.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 6.3 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.1 | 4.3 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.1 | 2.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 30.7 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.1 | 0.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 5.5 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 3.7 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 4.8 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 1.3 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 6.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 1.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 5.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 4.0 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 1.6 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 2.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 1.5 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 15.9 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 0.7 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 7.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 2.7 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.1 | 0.3 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.7 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 1.7 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.9 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.9 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 5.2 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.1 | 2.2 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.1 | 3.1 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 0.4 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.3 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 | 2.7 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 1.0 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.5 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 3.5 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.9 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 1.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 2.7 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 0.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 1.1 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.9 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 2.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.5 | GO:0090190 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.6 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.5 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 0.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 2.4 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 4.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 2.0 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 3.8 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 5.6 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.1 | 1.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.2 | GO:0051659 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 3.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 2.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 1.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 2.6 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 1.7 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 | 0.1 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 3.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.8 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 5.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 1.2 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 4.5 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 1.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 2.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.6 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 2.7 | GO:0098869 | cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 2.0 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 1.7 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.3 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 1.0 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 2.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 3.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 3.4 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.5 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.3 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.4 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 1.2 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 3.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.8 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 1.8 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 1.1 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.9 | 206.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 4.8 | 43.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 4.4 | 21.9 | GO:0036128 | CatSper complex(GO:0036128) |
| 2.8 | 8.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.5 | 29.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 2.3 | 6.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 2.0 | 8.0 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 2.0 | 27.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.8 | 18.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.8 | 8.9 | GO:0045160 | myosin I complex(GO:0045160) |
| 1.6 | 10.9 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 1.5 | 22.9 | GO:0030478 | actin cap(GO:0030478) |
| 1.5 | 17.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 1.4 | 7.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 1.3 | 26.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 1.2 | 3.6 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 1.1 | 8.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.0 | 17.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.9 | 2.7 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.9 | 4.4 | GO:0031673 | H zone(GO:0031673) |
| 0.9 | 14.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.8 | 12.6 | GO:0097433 | dense body(GO:0097433) |
| 0.8 | 4.9 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.8 | 17.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.7 | 25.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.6 | 5.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.6 | 2.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.6 | 5.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.5 | 2.1 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.5 | 1.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.5 | 7.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.5 | 21.1 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.5 | 1.4 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.4 | 4.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.4 | 2.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 2.9 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.4 | 3.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.4 | 4.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.4 | 2.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.4 | 4.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.4 | 2.4 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.4 | 2.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.4 | 41.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.4 | 45.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.4 | 14.8 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 2.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.3 | 2.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 9.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.3 | 5.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.3 | 13.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 2.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.3 | 6.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 10.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 5.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 2.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 2.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 7.3 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.2 | 22.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 2.9 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 3.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 11.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 11.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 7.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 5.6 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.2 | 6.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 1.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 8.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 19.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 3.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 6.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 38.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 12.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 13.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 2.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 4.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 1.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 18.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.9 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 11.7 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.3 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.1 | 1.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 4.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 2.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 5.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 23.9 | GO:0044432 | endoplasmic reticulum part(GO:0044432) |
| 0.1 | 1.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 3.5 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 13.7 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 1.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 8.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 14.7 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.5 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 10.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 21.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 23.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 1.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.4 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 11.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 5.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 4.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 5.0 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 1.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 8.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 5.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 5.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.1 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.6 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 1.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 4.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 34.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 25.1 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 22.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 3.3 | 13.3 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 3.0 | 11.9 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 2.3 | 6.8 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 2.3 | 15.8 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 2.1 | 25.1 | GO:0008430 | selenium binding(GO:0008430) |
| 2.0 | 179.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 1.8 | 5.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.7 | 10.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.7 | 9.9 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 1.7 | 6.6 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 1.6 | 8.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 1.5 | 4.5 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 1.4 | 7.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.4 | 5.8 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 1.4 | 8.6 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 1.4 | 5.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.4 | 9.6 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 1.3 | 5.3 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 1.3 | 12.8 | GO:0042731 | PH domain binding(GO:0042731) |
| 1.3 | 16.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.2 | 3.6 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 1.1 | 3.4 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 1.1 | 28.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 1.1 | 15.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.1 | 14.7 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 1.0 | 21.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.0 | 10.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 1.0 | 2.9 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.0 | 13.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.9 | 6.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.8 | 4.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.8 | 7.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.8 | 2.4 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.8 | 6.2 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.8 | 5.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.7 | 8.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.7 | 14.2 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.7 | 10.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.7 | 2.2 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.7 | 2.9 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.7 | 2.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.7 | 2.1 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.7 | 4.8 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.7 | 4.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.7 | 7.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.7 | 70.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.7 | 3.3 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.6 | 3.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.6 | 2.6 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.6 | 2.6 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.6 | 8.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.6 | 4.4 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.6 | 5.0 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.6 | 5.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.6 | 1.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.6 | 3.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.6 | 2.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.6 | 2.3 | GO:0008513 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.6 | 2.9 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.6 | 19.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.6 | 6.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.6 | 7.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.5 | 16.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.5 | 3.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.5 | 3.8 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.5 | 3.7 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.5 | 14.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.5 | 2.4 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.5 | 1.4 | GO:0042806 | fucose binding(GO:0042806) |
| 0.5 | 1.8 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.4 | 4.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.4 | 1.8 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.4 | 13.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 8.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.4 | 42.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 10.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 3.3 | GO:0005534 | galactose binding(GO:0005534) |
| 0.4 | 3.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.4 | 9.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.4 | 3.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.4 | 9.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 5.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.4 | 1.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.4 | 1.9 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.4 | 0.7 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.4 | 2.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.4 | 2.2 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.4 | 6.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 12.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 5.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.4 | 5.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 1.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 1.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.3 | 7.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 8.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 1.6 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.3 | 4.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.3 | 2.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 5.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.3 | 4.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 2.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 16.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 1.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.3 | 0.6 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.3 | 16.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 17.1 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
| 0.3 | 2.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.3 | 0.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.3 | 2.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 1.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 1.0 | GO:0032560 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) guanyl deoxyribonucleotide binding(GO:0032560) dGTP binding(GO:0032567) |
| 0.3 | 2.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.3 | 1.6 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.3 | 0.8 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.3 | 3.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.3 | 5.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.3 | 2.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.3 | 3.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.3 | 5.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 6.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 34.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 3.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 7.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.2 | 3.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 8.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 1.8 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.2 | 7.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 14.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 11.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 3.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 0.8 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.2 | 1.5 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 1.7 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 0.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 4.7 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.2 | 1.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 1.6 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 3.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 0.9 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 3.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 2.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 11.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 0.7 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.2 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 1.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 1.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.2 | 1.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 1.5 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 1.5 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 5.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 0.9 | GO:0050294 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 1.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 27.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 37.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.0 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 0.8 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 13.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.4 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 3.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 3.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.3 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 5.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 1.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 3.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 2.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.4 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 2.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 2.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.1 | 4.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.1 | GO:0070008 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 3.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 4.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.2 | GO:0030290 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.1 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 5.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 4.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 2.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 2.4 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.0 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 2.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 11.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 4.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 4.8 | GO:0016597 | amino acid binding(GO:0016597) |
| 0.0 | 4.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 11.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 2.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 2.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 4.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 8.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.5 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 4.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 6.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 10.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 1.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.4 | GO:0019210 | kinase inhibitor activity(GO:0019210) |
| 0.0 | 11.1 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 5.2 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 164.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 1.6 | 75.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.6 | 48.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.5 | 5.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.4 | 72.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.4 | 11.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 10.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.4 | 7.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.4 | 17.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.3 | 24.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 18.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.3 | 3.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.3 | 3.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.3 | 12.8 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.3 | 5.9 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.3 | 2.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.3 | 7.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.3 | 10.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 31.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 66.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 1.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 2.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 10.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.2 | 35.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 3.3 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.2 | 6.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 7.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 14.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 8.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 6.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.2 | 1.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 1.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 4.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 2.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 1.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 0.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 3.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 2.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 2.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 157.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 2.5 | 5.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 1.4 | 94.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 1.1 | 12.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 1.0 | 22.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 1.0 | 6.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.8 | 14.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.8 | 10.9 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.7 | 17.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.7 | 9.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.7 | 37.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.6 | 27.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.6 | 22.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.5 | 25.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.5 | 10.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.5 | 12.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.4 | 2.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 9.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.4 | 3.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.3 | 8.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 5.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.3 | 10.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.3 | 3.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 10.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 3.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 5.8 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.2 | 2.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 8.7 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.2 | 3.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 4.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 2.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 3.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 3.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 3.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 6.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 11.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 8.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 22.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.2 | 2.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 3.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 3.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 6.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 7.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 3.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.2 | 1.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 3.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 1.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 5.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 3.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 14.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.0 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 2.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.8 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 3.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 2.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 3.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 6.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 3.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 1.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 0.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.6 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 4.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 2.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 11.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 0.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 0.8 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.6 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 10.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.1 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.1 | 1.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.7 | REACTOME SHC MEDIATED CASCADE | Genes involved in SHC-mediated cascade |
| 0.0 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 2.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.1 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 1.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 3.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |