Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
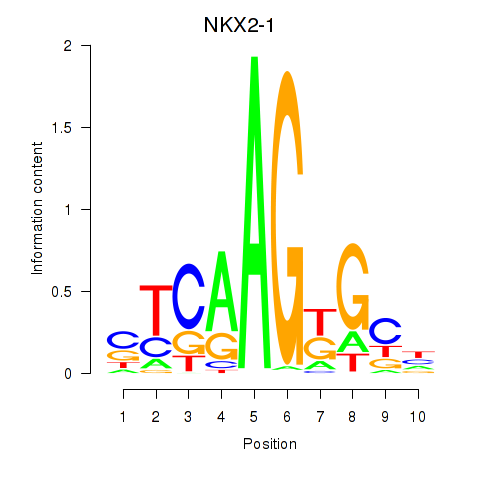
Results for NKX2-1
Z-value: 1.79
Transcription factors associated with NKX2-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-1
|
ENSG00000136352.19 | NKX2-1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-1 | hg38_v1_chr14_-_36521149_36521191, hg38_v1_chr14_-_36520222_36520237, hg38_v1_chr14_-_36519679_36519693 | 0.13 | 6.2e-02 | Click! |
Activity profile of NKX2-1 motif
Sorted Z-values of NKX2-1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 12.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 1.3 | 4.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 1.3 | 5.2 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 1.2 | 6.2 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 1.2 | 6.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.2 | 12.0 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 1.1 | 3.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.1 | 4.4 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.8 | 3.2 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.7 | 2.2 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.7 | 2.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.7 | 3.4 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.6 | 11.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.6 | 1.8 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.4 | 1.6 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.3 | 1.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 1.3 | GO:1904588 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.3 | 4.0 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.3 | 1.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.3 | 0.9 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.3 | 0.9 | GO:2000374 | regulation of oxygen metabolic process(GO:2000374) |
| 0.3 | 4.3 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.3 | 1.3 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.3 | 2.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.2 | 4.2 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.2 | 2.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 1.5 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.2 | 2.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 1.5 | GO:0072719 | copper ion import(GO:0015677) cellular response to cisplatin(GO:0072719) |
| 0.2 | 1.4 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.2 | 0.8 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 1.7 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.2 | 2.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.1 | GO:0021502 | neural fold elevation formation(GO:0021502) allantois development(GO:1905069) |
| 0.1 | 1.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 1.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 2.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 1.2 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 2.1 | GO:0003334 | keratinocyte development(GO:0003334) cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.6 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.1 | 3.2 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 1.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 1.3 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.3 | GO:0070662 | erythropoietin-mediated signaling pathway(GO:0038162) mast cell proliferation(GO:0070662) |
| 0.1 | 3.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 2.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 1.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 2.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.7 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.7 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 3.9 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.1 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.0 | 0.6 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of activated T cell proliferation(GO:0046007) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 4.6 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 2.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 6.4 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 3.9 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:1904504 | lipophagy(GO:0061724) regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 1.7 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.8 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 2.8 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 1.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.9 | GO:0070988 | demethylation(GO:0070988) |
| 0.0 | 3.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 1.3 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 1.0 | GO:0051437 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) positive regulation of ubiquitin-protein ligase activity involved in regulation of mitotic cell cycle transition(GO:0051437) |
| 0.0 | 0.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 1.5 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.7 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.2 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.4 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 1.8 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.8 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 1.0 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 12.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.8 | 12.0 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.5 | 6.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.5 | 4.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.5 | 5.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.4 | 4.6 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.4 | 3.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 4.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 4.4 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.3 | 1.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 2.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 4.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.3 | 6.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 2.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 1.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 3.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 0.9 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.4 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 2.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 3.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 5.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.0 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 2.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 4.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 3.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 2.1 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 15.3 | GO:0045121 | membrane raft(GO:0045121) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 3.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 5.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 1.6 | 11.0 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 1.1 | 3.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.8 | 3.2 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.7 | 3.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.7 | 2.2 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.6 | 3.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.5 | 1.6 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.4 | 8.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.4 | 1.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 2.3 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 3.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 0.7 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 5.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.2 | 2.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 1.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 1.3 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.2 | 4.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 1.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 16.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 2.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 2.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 2.2 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 1.6 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 2.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 4.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 1.9 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 2.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 3.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 2.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 3.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.2 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 4.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 5.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 3.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 5.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.5 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 4.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 5.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 12.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 11.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.3 | 6.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 7.2 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.2 | 2.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.2 | 12.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 4.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 2.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 3.9 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 2.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 4.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.9 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 2.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 4.4 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 1.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 4.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 3.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 2.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |