Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
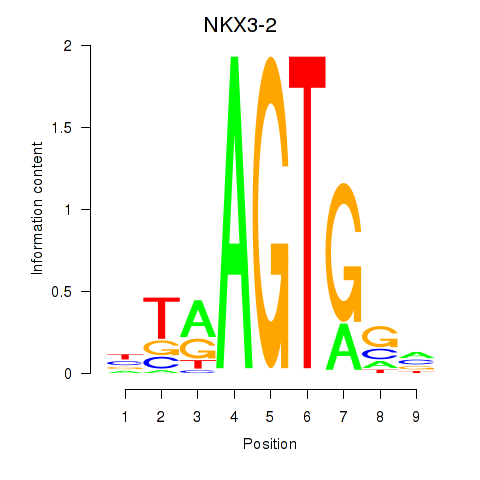
Results for NKX3-2
Z-value: 4.34
Transcription factors associated with NKX3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX3-2
|
ENSG00000109705.8 | NKX3-2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX3-2 | hg38_v1_chr4_-_13544506_13544513 | 0.19 | 5.6e-03 | Click! |
Activity profile of NKX3-2 motif
Sorted Z-values of NKX3-2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX3-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 20.2 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 5.9 | 23.6 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 3.3 | 10.0 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 3.1 | 9.2 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 1.6 | 6.3 | GO:1902617 | response to fluoride(GO:1902617) |
| 1.5 | 4.4 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 1.3 | 6.4 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 1.2 | 3.7 | GO:0015847 | putrescine transport(GO:0015847) |
| 1.1 | 5.7 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 1.1 | 3.4 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 1.1 | 4.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.0 | 3.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 1.0 | 9.4 | GO:0070543 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 1.0 | 3.0 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 1.0 | 66.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.9 | 10.0 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.9 | 3.5 | GO:1905069 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme morphogenesis(GO:0072134) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) allantois development(GO:1905069) |
| 0.8 | 5.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.8 | 4.6 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.7 | 2.0 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.6 | 5.0 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.6 | 4.7 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.6 | 2.3 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.6 | 2.3 | GO:0098923 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.6 | 6.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.6 | 2.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.5 | 1.6 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.5 | 2.0 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.5 | 7.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.5 | 2.5 | GO:0030242 | pexophagy(GO:0030242) aggrephagy(GO:0035973) |
| 0.5 | 1.5 | GO:0006683 | galactosylceramide catabolic process(GO:0006683) |
| 0.5 | 3.2 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.4 | 4.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.4 | 1.3 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) trochlear nerve development(GO:0021558) regulation of timing of neuron differentiation(GO:0060164) |
| 0.4 | 2.6 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.4 | 5.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.4 | 1.3 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.4 | 3.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.4 | 4.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.4 | 1.3 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.4 | 6.7 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.4 | 7.6 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.4 | 2.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.4 | 8.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.4 | 2.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.4 | 7.4 | GO:0015671 | gas transport(GO:0015669) oxygen transport(GO:0015671) |
| 0.4 | 4.2 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.4 | 1.4 | GO:0048850 | hypophysis morphogenesis(GO:0048850) diencephalon morphogenesis(GO:0048852) |
| 0.4 | 3.9 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.4 | 3.2 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.3 | 1.0 | GO:0002668 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.3 | 1.7 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.3 | 9.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 7.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 15.0 | GO:0033198 | response to ATP(GO:0033198) |
| 0.3 | 1.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.3 | 2.6 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.3 | 1.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.3 | 8.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.3 | 1.5 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.3 | 3.8 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.3 | 2.0 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.3 | 0.8 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.3 | 2.4 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.3 | 0.8 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.3 | 0.8 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.3 | 5.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.3 | 3.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.3 | 1.6 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.3 | 1.0 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.3 | 1.5 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.3 | 2.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 1.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 1.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 5.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 9.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 8.5 | GO:0007099 | centriole replication(GO:0007099) centriole assembly(GO:0098534) |
| 0.2 | 20.1 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.2 | 4.3 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.2 | 12.1 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.2 | 31.0 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.2 | 2.5 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.2 | 13.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 1.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 2.6 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.2 | 1.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) activation of phospholipase A2 activity(GO:0032431) |
| 0.2 | 13.1 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.2 | 0.7 | GO:0086053 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.2 | 5.7 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 0.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 4.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 1.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 2.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.2 | 0.8 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 2.0 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.2 | 3.2 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.2 | 1.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 1.9 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.1 | 3.0 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.1 | 0.4 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 1.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 1.0 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 1.7 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 2.1 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 2.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 7.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.7 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 2.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 1.7 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.1 | 1.8 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 1.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 9.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 3.0 | GO:0016246 | RNA interference(GO:0016246) |
| 0.1 | 4.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.7 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.1 | 1.6 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 1.9 | GO:0002438 | acute inflammatory response to antigenic stimulus(GO:0002438) |
| 0.1 | 2.1 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.5 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 2.1 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 1.7 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.1 | 0.3 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.1 | 6.0 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.1 | 2.3 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 1.9 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 1.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 1.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.9 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.6 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 2.7 | GO:0006506 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 2.3 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 1.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.4 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 1.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 2.1 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 1.5 | GO:0048026 | positive regulation of circadian rhythm(GO:0042753) positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 3.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.6 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 0.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 4.4 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.1 | 4.5 | GO:0048477 | oogenesis(GO:0048477) |
| 0.1 | 1.1 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.1 | 6.3 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.1 | 0.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 2.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.7 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 3.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.1 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 3.0 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 5.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.8 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 2.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.3 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.5 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 2.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.4 | GO:0070327 | lysine catabolic process(GO:0006554) thyroid hormone transport(GO:0070327) |
| 0.0 | 12.2 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 1.2 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 1.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.3 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 3.0 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 2.7 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 3.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 2.6 | GO:0048705 | skeletal system morphogenesis(GO:0048705) |
| 0.0 | 0.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 3.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 1.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 4.2 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.7 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.0 | 1.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.0 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 2.1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.6 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.9 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 57.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.8 | 9.2 | GO:0042825 | TAP complex(GO:0042825) |
| 1.6 | 8.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.5 | 10.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 1.4 | 8.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 1.3 | 66.5 | GO:0042571 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 1.0 | 7.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.9 | 7.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.8 | 8.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.8 | 5.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.8 | 6.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.8 | 6.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.7 | 13.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.7 | 5.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.5 | 7.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.5 | 2.3 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.4 | 3.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.4 | 6.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.4 | 1.5 | GO:0035363 | histone locus body(GO:0035363) |
| 0.4 | 4.4 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.3 | 1.4 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.3 | 13.9 | GO:0000791 | euchromatin(GO:0000791) |
| 0.3 | 5.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.3 | 5.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 0.6 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.3 | 14.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 2.0 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.3 | 1.9 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.3 | 0.8 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.3 | 1.6 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.3 | 2.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.3 | 9.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 3.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.2 | 1.9 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.2 | 6.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 5.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 1.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 3.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 0.5 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.2 | 4.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 2.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.8 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 2.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 2.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 4.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 2.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 2.0 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 2.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 4.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 13.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 7.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 2.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 3.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 2.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 3.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.2 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.1 | 1.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 3.1 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 5.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 2.1 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.6 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 1.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.1 | GO:0036020 | AP-2 adaptor complex(GO:0030122) endolysosome membrane(GO:0036020) |
| 0.0 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 3.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 5.1 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 4.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 11.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 2.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 21.7 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 3.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 2.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 2.4 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.8 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.4 | GO:0044438 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 1.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 20.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 2.5 | 10.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 2.0 | 10.0 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 1.7 | 10.4 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 1.5 | 9.2 | GO:0046979 | peptide-transporting ATPase activity(GO:0015440) TAP2 binding(GO:0046979) |
| 1.5 | 7.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 1.5 | 13.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 1.5 | 24.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 1.4 | 35.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 1.3 | 6.7 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 1.2 | 3.7 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 1.2 | 9.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 1.2 | 3.5 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 1.1 | 3.4 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 1.1 | 7.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 1.1 | 66.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.9 | 4.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.9 | 2.6 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.8 | 2.4 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.7 | 4.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.7 | 2.0 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.7 | 2.0 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.6 | 5.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.6 | 1.7 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.5 | 3.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.5 | 2.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.5 | 1.5 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.5 | 7.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.5 | 1.9 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.5 | 13.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.5 | 4.2 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.4 | 2.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.4 | 5.7 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.4 | 6.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.4 | 5.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.4 | 2.3 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.4 | 3.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 2.6 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.4 | 2.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.4 | 1.9 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.4 | 0.7 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.3 | 3.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 4.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 3.3 | GO:0001727 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 9.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 2.2 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.3 | 1.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 1.6 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.3 | 5.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.3 | 3.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.3 | 3.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.3 | 4.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 3.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 1.0 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 1.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 6.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 2.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 2.4 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 3.9 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 2.7 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 0.9 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.2 | 1.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.2 | 1.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.2 | 3.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 0.8 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 5.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 4.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 0.6 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.2 | 2.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 1.4 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.2 | 3.0 | GO:0016594 | glycine binding(GO:0016594) |
| 0.2 | 2.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 2.6 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 1.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 3.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 2.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 0.8 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 3.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 10.8 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 1.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 2.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 1.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.4 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 2.8 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 2.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.0 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 3.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.8 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 2.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 3.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 12.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.6 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 4.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 9.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 20.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 27.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.0 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.1 | 0.9 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 1.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 1.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.7 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 1.0 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 1.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 2.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 2.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 1.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 2.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 2.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.5 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 7.2 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.3 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 1.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 2.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 1.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 4.2 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 2.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.9 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 2.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 11.8 | GO:0004888 | transmembrane signaling receptor activity(GO:0004888) |
| 0.0 | 0.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 27.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.5 | 13.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.4 | 23.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.4 | 6.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 9.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 6.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 8.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 8.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 9.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 8.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 20.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 3.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 3.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 4.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 8.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 5.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 9.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 4.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 6.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 0.7 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 2.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 10.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 5.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 23.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.7 | 24.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.4 | 8.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 3.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.3 | 4.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 16.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.3 | 5.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.3 | 8.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.3 | 10.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 4.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 8.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 5.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 6.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 4.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 20.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.2 | 6.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 3.9 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.2 | 7.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 8.6 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.1 | 2.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 7.9 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 8.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 4.7 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.1 | 1.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 2.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 3.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 2.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 9.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.9 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.6 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 1.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 2.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 4.9 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.1 | 3.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 3.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.5 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 1.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 3.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 4.7 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 3.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 6.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 2.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 3.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |