Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
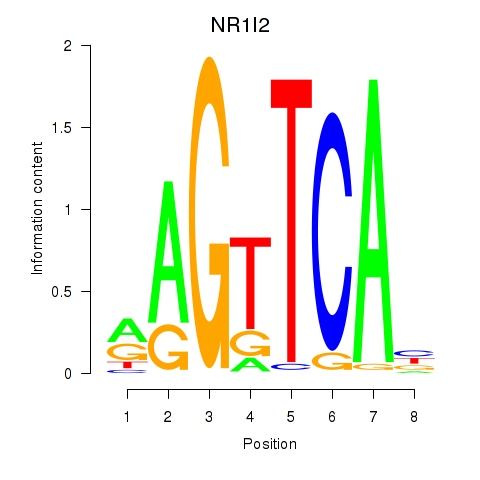
Results for NR1I2
Z-value: 3.12
Transcription factors associated with NR1I2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I2
|
ENSG00000144852.20 | NR1I2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1I2 | hg38_v1_chr3_+_119782094_119782114 | 0.25 | 1.4e-04 | Click! |
Activity profile of NR1I2 motif
Sorted Z-values of NR1I2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 78.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 4.6 | 13.9 | GO:0060875 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 4.2 | 12.6 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 3.1 | 12.3 | GO:0009956 | radial pattern formation(GO:0009956) |
| 3.0 | 14.9 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 2.6 | 10.5 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 2.6 | 7.8 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 2.5 | 7.4 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 2.4 | 7.3 | GO:0015847 | putrescine transport(GO:0015847) |
| 2.0 | 8.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 1.8 | 7.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 1.6 | 6.2 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 1.5 | 4.5 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 1.5 | 25.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.4 | 12.8 | GO:1902037 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 1.3 | 10.4 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 1.3 | 3.9 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 1.3 | 6.4 | GO:0001555 | oocyte growth(GO:0001555) |
| 1.2 | 12.5 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 1.2 | 4.8 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 1.2 | 3.5 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 1.2 | 7.1 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 1.1 | 12.4 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.1 | 4.5 | GO:0035054 | atrial septum secundum morphogenesis(GO:0003290) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 1.1 | 3.3 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 1.1 | 5.5 | GO:0090131 | skeletal muscle thin filament assembly(GO:0030240) mesenchyme migration(GO:0090131) |
| 1.1 | 4.3 | GO:0036378 | alkaloid catabolic process(GO:0009822) calcitriol biosynthetic process from calciol(GO:0036378) |
| 1.1 | 2.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 1.1 | 9.5 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 1.1 | 3.2 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 1.0 | 3.8 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.9 | 12.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.9 | 8.4 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.9 | 26.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.9 | 2.7 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.9 | 31.5 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.9 | 5.2 | GO:0003051 | angiotensin catabolic process in blood(GO:0002005) angiotensin-mediated drinking behavior(GO:0003051) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) |
| 0.8 | 3.4 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.8 | 2.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.8 | 4.7 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.8 | 3.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.8 | 2.3 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.7 | 4.5 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.7 | 12.4 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.7 | 4.3 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.7 | 2.0 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.7 | 2.0 | GO:0060003 | copper ion export(GO:0060003) |
| 0.7 | 13.2 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.6 | 1.8 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.6 | 5.4 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.6 | 1.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.6 | 1.8 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.5 | 1.6 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.5 | 1.6 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.5 | 2.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.5 | 3.7 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.5 | 3.7 | GO:0016045 | detection of bacterium(GO:0016045) |
| 0.5 | 4.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.5 | 6.6 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.5 | 17.4 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.5 | 2.4 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.5 | 5.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.5 | 4.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.4 | 6.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.4 | 5.8 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.4 | 4.8 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.4 | 3.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.4 | 5.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.4 | 27.7 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.4 | 1.4 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.4 | 4.6 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.4 | 1.8 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) regulation of stem cell division(GO:2000035) |
| 0.4 | 1.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.3 | 4.4 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.3 | 3.0 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.3 | 3.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.3 | 3.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.3 | 10.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.3 | 2.6 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.3 | 3.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.3 | 2.9 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.3 | 3.1 | GO:0014826 | artery smooth muscle contraction(GO:0014824) vein smooth muscle contraction(GO:0014826) |
| 0.3 | 5.4 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.3 | 4.2 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.3 | 28.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.3 | 14.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.3 | 7.8 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.3 | 2.7 | GO:0046959 | habituation(GO:0046959) |
| 0.3 | 1.6 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.3 | 1.8 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.3 | 3.8 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 1.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.2 | 4.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 | 1.2 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.2 | 1.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 6.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 6.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 1.5 | GO:0060413 | atrial septum morphogenesis(GO:0060413) |
| 0.2 | 5.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 0.6 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.2 | 3.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 0.6 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.2 | 0.8 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.2 | 3.4 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.2 | 1.7 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.2 | 6.7 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 1.8 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.2 | 3.9 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 3.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 9.9 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.2 | 2.3 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 1.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 3.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 5.4 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.2 | 2.7 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.2 | 3.9 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 3.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 1.5 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 1.0 | GO:0071692 | sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.1 | 1.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 2.9 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.7 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 1.7 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.1 | 2.4 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.1 | 2.3 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 2.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 1.4 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 18.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 1.1 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.1 | 1.0 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 1.2 | GO:1901977 | negative regulation of cell cycle checkpoint(GO:1901977) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 0.8 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 1.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.8 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 2.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 2.3 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 0.5 | GO:0032764 | smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 1.6 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.1 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 2.7 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 | 0.2 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.4 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 4.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 0.5 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.6 | GO:0009595 | detection of biotic stimulus(GO:0009595) |
| 0.1 | 7.8 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 9.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.8 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.1 | 1.0 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 2.7 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 4.0 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.5 | GO:0051583 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.1 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 9.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 4.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 0.4 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 2.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.5 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.1 | 1.5 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.2 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 2.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 1.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.7 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 6.7 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 2.1 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 1.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 4.0 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.1 | 4.6 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.1 | 3.2 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.1 | 2.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.4 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.9 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 2.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 1.5 | GO:0060393 | regulation of pathway-restricted SMAD protein phosphorylation(GO:0060393) |
| 0.0 | 5.6 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 6.0 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 3.5 | GO:0060996 | dendritic spine development(GO:0060996) |
| 0.0 | 3.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 2.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 6.9 | GO:0048839 | inner ear development(GO:0048839) |
| 0.0 | 2.1 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 1.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 5.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 5.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 2.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.7 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.9 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 5.6 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 3.7 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 1.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 2.4 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 2.5 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.0 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.9 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 2.7 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 3.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 2.7 | GO:0007601 | visual perception(GO:0007601) sensory perception of light stimulus(GO:0050953) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.0 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.4 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 67.9 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.8 | 28.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.6 | 4.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 1.2 | 11.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.9 | 4.7 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.9 | 10.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.7 | 4.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.6 | 12.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.5 | 6.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 12.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.5 | 16.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.5 | 6.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 3.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.4 | 2.9 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.3 | 13.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.3 | 1.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.3 | 2.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.3 | 1.5 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 4.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 8.2 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.3 | 4.3 | GO:0001741 | XY body(GO:0001741) |
| 0.3 | 3.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 0.8 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 1.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 2.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 6.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 4.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 4.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 21.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 3.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 2.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 2.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 5.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 2.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 1.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 1.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 5.4 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.2 | 2.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 5.5 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 1.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 2.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 5.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.3 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 5.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 10.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.7 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 10.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 13.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 3.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 3.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 10.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 2.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 22.8 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 5.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 5.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 2.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 16.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 4.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 13.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 3.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 2.9 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.1 | 4.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 3.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 36.9 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.1 | 3.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 1.0 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 7.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 3.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 19.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 6.9 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 4.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 50.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 4.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 3.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 13.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 2.4 | 7.3 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 2.1 | 10.4 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 2.1 | 6.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 2.0 | 8.1 | GO:0035473 | lipase binding(GO:0035473) |
| 1.7 | 17.3 | GO:0005549 | odorant binding(GO:0005549) |
| 1.6 | 4.7 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 1.4 | 4.3 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 1.4 | 7.2 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 1.1 | 4.5 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 1.0 | 5.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 1.0 | 3.9 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.9 | 5.4 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.9 | 14.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.8 | 24.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.8 | 69.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.8 | 2.4 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.8 | 4.6 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.7 | 4.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.7 | 4.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.7 | 2.0 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.7 | 4.7 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.6 | 3.8 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.6 | 3.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.6 | 1.8 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.6 | 3.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.6 | 5.8 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.6 | 6.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.6 | 3.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.6 | 1.7 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.6 | 3.4 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.5 | 6.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.5 | 3.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.5 | 3.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.5 | 1.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.5 | 3.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.5 | 1.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.5 | 2.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.4 | 3.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.4 | 4.8 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.4 | 8.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.4 | 4.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.4 | 1.5 | GO:0005334 | norepinephrine:sodium symporter activity(GO:0005334) |
| 0.4 | 5.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.4 | 14.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.4 | 4.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.4 | 4.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.4 | 28.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 1.7 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.3 | 4.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.3 | 1.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 2.6 | GO:0005534 | galactose binding(GO:0005534) |
| 0.3 | 6.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 7.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.3 | 1.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.3 | 2.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 5.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 12.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.3 | 3.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 3.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 3.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 2.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 13.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 1.4 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.2 | 0.9 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.2 | 1.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 2.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 1.3 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.2 | 12.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 1.7 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 2.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 4.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 1.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 7.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 2.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 2.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 10.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 2.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 6.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 1.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 2.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 4.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 3.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 0.9 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.2 | 3.9 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 2.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 6.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 6.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 4.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 5.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 1.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 1.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 4.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 2.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 4.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 2.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 11.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.7 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 1.6 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 6.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 3.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 9.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 1.8 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 2.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 12.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 17.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 11.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 6.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 2.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.5 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 20.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 4.2 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.1 | 9.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 29.1 | GO:0001159 | core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 16.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 3.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.2 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 29.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 2.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 2.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 3.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 5.6 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 1.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 9.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 2.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 4.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.8 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 2.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 9.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 2.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 7.5 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 64.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.5 | 14.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.4 | 26.1 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.3 | 27.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 13.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 6.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 16.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 23.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 4.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 2.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 8.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 5.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 6.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 6.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 13.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 37.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 9.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 2.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 3.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 6.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 4.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 2.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 5.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 3.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 67.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.6 | 6.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.5 | 6.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.5 | 13.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.4 | 11.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.4 | 12.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.4 | 9.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.4 | 19.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.4 | 12.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 28.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.3 | 3.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 4.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 4.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 4.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 26.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 3.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 4.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 5.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 1.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 3.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 6.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 23.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 3.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 1.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 3.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 20.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 2.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 21.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 20.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 3.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 3.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 5.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.7 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 6.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 4.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 13.6 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 5.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 3.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 3.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 2.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 3.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 2.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 4.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.8 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 2.0 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.1 | 1.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 4.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 2.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 5.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.7 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |