Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
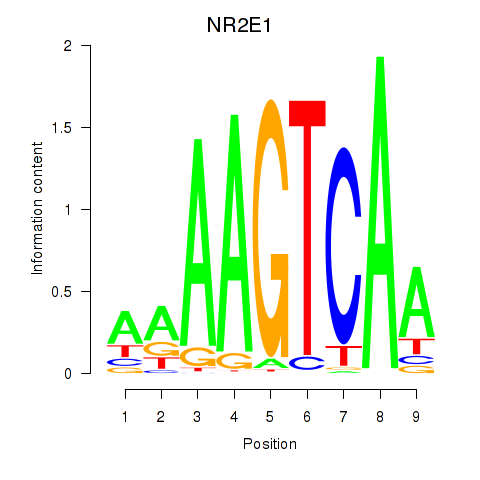
Results for NR2E1
Z-value: 2.36
Transcription factors associated with NR2E1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E1
|
ENSG00000112333.12 | NR2E1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2E1 | hg38_v1_chr6_+_108166015_108166034 | 0.31 | 3.2e-06 | Click! |
Activity profile of NR2E1 motif
Sorted Z-values of NR2E1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 39.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 2.3 | 9.3 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 2.1 | 23.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.9 | 26.9 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 1.7 | 6.8 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 1.7 | 5.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.7 | 5.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 1.7 | 6.7 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 1.6 | 4.9 | GO:0022018 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 1.5 | 4.5 | GO:2000048 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 1.4 | 4.1 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 1.2 | 16.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 1.2 | 3.7 | GO:0019046 | release from viral latency(GO:0019046) |
| 1.2 | 23.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.1 | 9.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.1 | 22.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.1 | 3.2 | GO:0021592 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 1.0 | 8.4 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 1.0 | 16.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 1.0 | 5.8 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.9 | 5.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.9 | 5.2 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.9 | 3.4 | GO:0035054 | atrial septum secundum morphogenesis(GO:0003290) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.8 | 5.8 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.8 | 2.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.8 | 10.5 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.8 | 10.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.7 | 5.6 | GO:0035989 | tendon development(GO:0035989) |
| 0.7 | 3.5 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.7 | 4.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.7 | 2.7 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.7 | 8.0 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.7 | 3.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.6 | 2.5 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.6 | 1.9 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.6 | 2.5 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.6 | 3.6 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.5 | 2.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.5 | 2.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.5 | 3.7 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.5 | 1.6 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.5 | 5.6 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.5 | 2.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.5 | 2.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.5 | 3.4 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.5 | 12.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.5 | 4.6 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.5 | 2.3 | GO:0097491 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.5 | 2.7 | GO:0060615 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) mammary gland specification(GO:0060594) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.4 | 1.3 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.4 | 1.3 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.4 | 3.6 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.4 | 1.2 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.4 | 5.9 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.4 | 4.7 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.4 | 1.9 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.4 | 6.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.4 | 19.5 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.4 | 9.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.4 | 9.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.4 | 1.8 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.4 | 1.4 | GO:0060529 | squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.4 | 2.8 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.3 | 1.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.3 | 7.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 1.0 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.3 | 2.5 | GO:0001554 | luteolysis(GO:0001554) |
| 0.3 | 1.8 | GO:0070171 | negative regulation of odontogenesis(GO:0042483) negative regulation of tooth mineralization(GO:0070171) |
| 0.3 | 1.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.3 | 2.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 0.8 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) negative regulation of icosanoid secretion(GO:0032304) |
| 0.3 | 2.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.3 | 0.5 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.3 | 1.0 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.2 | 2.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 1.7 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.2 | 5.0 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.2 | 8.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 11.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 5.9 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.2 | 5.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 5.6 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.2 | 2.7 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.2 | 1.2 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 2.2 | GO:0086068 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.2 | 2.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 3.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 3.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 3.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 4.1 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.2 | 1.3 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.2 | 3.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 1.6 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.2 | 1.8 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.2 | 3.3 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.2 | 6.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 5.3 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.2 | 3.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 4.2 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.2 | 2.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 2.1 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.2 | 0.3 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 3.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.6 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.1 | 1.0 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 2.9 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 2.6 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 1.2 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.1 | 10.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 2.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 1.8 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 1.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 3.9 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 5.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 11.8 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 2.1 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.1 | 2.3 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 0.9 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 4.0 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 0.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 1.0 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 1.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.3 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 2.0 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 2.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 6.7 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.1 | 4.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 2.1 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) |
| 0.1 | 3.3 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 2.8 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 1.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 3.3 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.6 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.0 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 1.6 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 2.6 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.1 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.2 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 1.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 1.4 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.8 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 4.1 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.2 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.1 | GO:0045162 | neuronal action potential propagation(GO:0019227) clustering of voltage-gated sodium channels(GO:0045162) action potential propagation(GO:0098870) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 39.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.9 | 5.6 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 1.7 | 21.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.6 | 23.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 1.3 | 5.3 | GO:1990879 | CST complex(GO:1990879) |
| 1.2 | 23.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 1.2 | 3.5 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.9 | 10.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.7 | 4.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.6 | 2.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.5 | 5.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.5 | 9.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.4 | 2.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.4 | 6.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.4 | 2.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.4 | 1.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 1.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 3.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 3.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.3 | 3.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 9.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 2.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 3.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 2.7 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.2 | 2.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 1.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 8.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 2.7 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.2 | 2.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 3.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 8.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.9 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 4.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.8 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 2.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 4.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 5.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 4.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 3.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 7.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 2.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.7 | GO:0034358 | plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.1 | 1.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 10.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 6.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.6 | GO:0098798 | mitochondrial protein complex(GO:0098798) |
| 0.1 | 3.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.5 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 1.3 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.1 | 7.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 3.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 5.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 9.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 5.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 41.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 3.6 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 3.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 2.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 5.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 23.4 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.8 | 16.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 1.7 | 6.8 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 1.6 | 23.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.4 | 41.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.2 | 8.4 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 1.1 | 5.7 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 1.0 | 9.8 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.9 | 2.8 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.9 | 10.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.9 | 3.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.8 | 2.3 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.8 | 9.0 | GO:0008430 | selenium binding(GO:0008430) |
| 0.7 | 5.0 | GO:0005497 | androgen binding(GO:0005497) |
| 0.6 | 10.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.6 | 4.5 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.5 | 4.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.5 | 2.5 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.5 | 1.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.5 | 5.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.5 | 3.7 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.5 | 2.8 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.5 | 1.8 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.4 | 2.7 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.4 | 10.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.4 | 3.5 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.4 | 7.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.4 | 1.3 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.4 | 2.5 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.4 | 1.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.4 | 32.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 5.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.4 | 3.7 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.4 | 12.0 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.4 | 3.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.3 | 3.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.3 | 3.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.3 | 3.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.3 | 3.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 3.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.3 | 1.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.3 | 5.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.3 | 7.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 5.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.2 | 5.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 2.9 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 1.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 3.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 2.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 1.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 1.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 13.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 5.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 2.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 6.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 0.9 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 0.8 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 2.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 3.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 2.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 3.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.9 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 6.4 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.1 | 0.9 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 2.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 1.0 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 3.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 1.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 2.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 5.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 5.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 2.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 1.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 3.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 5.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 9.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.9 | GO:0070492 | sialic acid binding(GO:0033691) oligosaccharide binding(GO:0070492) |
| 0.1 | 5.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 3.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 7.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.9 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 1.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 4.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 2.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 5.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 2.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 4.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 5.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 7.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 2.5 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 9.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 6.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.3 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 1.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 1.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 3.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 3.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.6 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 1.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 5.1 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 8.6 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 6.7 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 24.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.4 | 22.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 5.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 3.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 29.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 6.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 11.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 2.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 1.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 2.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 0.8 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 30.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 5.6 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.1 | 2.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 4.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 4.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 5.3 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 17.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 3.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 0.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 12.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 4.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 2.8 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 1.3 | 15.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.8 | 21.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.6 | 10.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.6 | 8.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.5 | 8.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 4.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 6.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.3 | 6.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 3.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 9.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 2.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 10.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 9.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 13.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 6.8 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.2 | 3.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 5.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.2 | 3.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 2.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 3.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 3.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 5.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 2.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.7 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 10.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 5.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 4.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 3.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 5.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 4.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 5.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 2.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 11.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 3.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 0.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 6.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 7.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 3.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.6 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.4 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |