Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
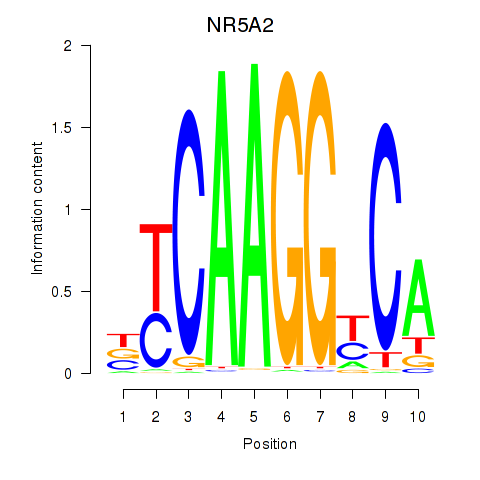
Results for NR5A2
Z-value: 7.04
Transcription factors associated with NR5A2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR5A2
|
ENSG00000116833.14 | NR5A2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR5A2 | hg38_v1_chr1_+_200027702_200027716 | 0.42 | 1.0e-10 | Click! |
Activity profile of NR5A2 motif
Sorted Z-values of NR5A2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR5A2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.8 | 320.3 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 12.4 | 37.3 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 9.2 | 55.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 8.1 | 32.5 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 6.8 | 20.3 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 5.9 | 23.5 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 5.7 | 17.0 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 5.5 | 16.6 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 5.4 | 16.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 5.3 | 31.7 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 5.0 | 34.8 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 4.9 | 14.7 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 4.9 | 24.3 | GO:1901909 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 4.6 | 13.9 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 4.5 | 36.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 3.7 | 11.1 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 3.7 | 18.3 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 3.6 | 14.6 | GO:0030035 | microspike assembly(GO:0030035) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 3.5 | 24.8 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 3.5 | 10.4 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 3.3 | 62.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 3.3 | 29.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 3.2 | 19.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 3.1 | 53.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 3.1 | 9.2 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 3.0 | 63.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 3.0 | 14.9 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 3.0 | 8.9 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 2.9 | 8.8 | GO:1902809 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 2.9 | 23.4 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 2.9 | 11.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 2.9 | 14.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 2.8 | 19.5 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 2.8 | 8.3 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 2.8 | 16.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 2.7 | 32.8 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 2.7 | 21.5 | GO:0061709 | reticulophagy(GO:0061709) |
| 2.7 | 50.6 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 2.6 | 49.8 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 2.6 | 10.5 | GO:0030242 | pexophagy(GO:0030242) |
| 2.6 | 13.0 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 2.6 | 10.2 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 2.5 | 53.0 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 2.5 | 7.6 | GO:0042323 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 2.5 | 7.5 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 2.5 | 9.8 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 2.5 | 7.4 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 2.4 | 14.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 2.3 | 6.8 | GO:0071529 | cementum mineralization(GO:0071529) |
| 2.2 | 6.6 | GO:1905237 | cellular response to mycotoxin(GO:0036146) response to antineoplastic agent(GO:0097327) response to cyclosporin A(GO:1905237) positive regulation of response to drug(GO:2001025) |
| 2.2 | 8.8 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 2.1 | 19.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 2.1 | 2.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 2.1 | 33.7 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 2.1 | 21.0 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 2.0 | 6.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 2.0 | 8.1 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 2.0 | 4.0 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 2.0 | 6.0 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 2.0 | 5.9 | GO:1905073 | uterine wall breakdown(GO:0042704) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 1.9 | 5.8 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 1.9 | 24.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 1.9 | 5.7 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 1.9 | 9.4 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 1.9 | 5.6 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 1.8 | 11.0 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 1.8 | 10.8 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 1.8 | 9.0 | GO:2000870 | oocyte growth(GO:0001555) regulation of progesterone secretion(GO:2000870) |
| 1.8 | 10.7 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 1.8 | 10.6 | GO:1904306 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 1.8 | 7.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 1.7 | 5.2 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 1.7 | 6.9 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 1.7 | 3.4 | GO:0046959 | habituation(GO:0046959) |
| 1.7 | 6.8 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 1.7 | 16.7 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.7 | 5.0 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 1.6 | 93.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 1.6 | 6.4 | GO:0007538 | primary sex determination(GO:0007538) |
| 1.6 | 7.9 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 1.5 | 7.7 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 1.5 | 4.6 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 1.5 | 15.1 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 1.5 | 7.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 1.4 | 4.3 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 1.4 | 5.8 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 1.4 | 5.8 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 1.4 | 4.1 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 1.4 | 15.0 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 1.4 | 9.6 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 1.4 | 4.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 1.3 | 14.8 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 1.3 | 6.7 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 1.3 | 5.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 1.3 | 7.8 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 1.3 | 7.7 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 1.3 | 22.9 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 1.2 | 18.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 1.2 | 9.6 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 1.2 | 3.6 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 1.2 | 17.9 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 1.2 | 4.8 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 1.2 | 7.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 1.2 | 7.0 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 1.1 | 9.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.1 | 5.7 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 1.1 | 9.0 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 1.1 | 15.7 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 1.1 | 8.7 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 1.1 | 2.2 | GO:0033058 | directional locomotion(GO:0033058) |
| 1.1 | 4.3 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 1.1 | 10.7 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 1.1 | 14.9 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.0 | 15.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.0 | 4.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 1.0 | 5.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 1.0 | 5.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 1.0 | 4.0 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 1.0 | 6.9 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 1.0 | 4.0 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 1.0 | 2.9 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 1.0 | 14.3 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.9 | 14.7 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.9 | 4.6 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.9 | 2.7 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.9 | 20.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.9 | 2.6 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.9 | 3.5 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.9 | 3.4 | GO:0052330 | induction of programmed cell death(GO:0012502) peptidyl-cysteine S-trans-nitrosylation(GO:0035606) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.8 | 15.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.8 | 5.8 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.8 | 9.9 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.8 | 3.3 | GO:1902938 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.8 | 4.1 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.8 | 19.5 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.8 | 8.9 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.8 | 3.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.8 | 17.4 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.8 | 8.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.8 | 7.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.7 | 9.7 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.7 | 20.1 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.7 | 6.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.7 | 2.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.7 | 2.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.7 | 7.6 | GO:0001787 | natural killer cell proliferation(GO:0001787) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.7 | 2.0 | GO:0044266 | angiotensin catabolic process in blood(GO:0002005) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) |
| 0.7 | 9.5 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.7 | 8.8 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.7 | 13.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.7 | 6.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.6 | 8.4 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.6 | 16.2 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.6 | 5.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.6 | 13.9 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.6 | 13.8 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.6 | 11.5 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.6 | 3.0 | GO:0071316 | olfactory nerve development(GO:0021553) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) cellular response to nicotine(GO:0071316) |
| 0.6 | 5.4 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.6 | 13.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.6 | 5.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.6 | 4.0 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.6 | 10.0 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.6 | 8.9 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.5 | 21.6 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.5 | 4.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.5 | 1.6 | GO:0060974 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.5 | 4.6 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.5 | 3.5 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.5 | 6.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.5 | 1.9 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.5 | 7.7 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.5 | 16.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.5 | 4.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.5 | 32.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.5 | 12.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.4 | 10.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.4 | 16.6 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.4 | 9.6 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.4 | 2.2 | GO:0051971 | response to L-ascorbic acid(GO:0033591) positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.4 | 1.3 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.4 | 5.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.4 | 11.2 | GO:0007618 | mating(GO:0007618) |
| 0.4 | 7.9 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.4 | 2.9 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.4 | 11.1 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.4 | 3.9 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.4 | 13.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.4 | 1.5 | GO:0072180 | pronephric nephron tubule development(GO:0039020) regulation of endodermal cell fate specification(GO:0042663) hepatoblast differentiation(GO:0061017) mesonephric duct morphogenesis(GO:0072180) |
| 0.4 | 1.5 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.4 | 5.6 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.4 | 28.0 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.4 | 1.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.4 | 3.0 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.4 | 6.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.4 | 4.4 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.4 | 5.0 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.4 | 13.4 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.4 | 3.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.4 | 7.7 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.3 | 2.7 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.3 | 6.2 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.3 | 0.7 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.3 | 7.1 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.3 | 9.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 5.3 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.3 | 1.3 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.3 | 18.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.3 | 2.6 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.3 | 3.5 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.3 | 1.9 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.3 | 10.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.3 | 6.0 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.3 | 20.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.3 | 2.6 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.3 | 0.3 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.3 | 1.7 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.3 | 3.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.3 | 0.8 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) lipopolysaccharide transport(GO:0015920) |
| 0.3 | 0.5 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.3 | 6.9 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.3 | 7.1 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.3 | 7.2 | GO:0060384 | innervation(GO:0060384) |
| 0.3 | 7.7 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.2 | 5.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 5.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.2 | 3.7 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.2 | 4.6 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.2 | 8.6 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.2 | 1.6 | GO:1902166 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.2 | 3.8 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.2 | 1.8 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.2 | 3.1 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.2 | 5.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 6.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 0.4 | GO:0071554 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.2 | 5.7 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.2 | 32.9 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.2 | 4.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 1.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 14.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.2 | 163.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.2 | 4.7 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.2 | 2.7 | GO:0032196 | transposition(GO:0032196) |
| 0.2 | 0.7 | GO:0098711 | iron ion import into cell(GO:0097459) iron ion import across plasma membrane(GO:0098711) |
| 0.2 | 11.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.2 | 10.0 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.2 | 6.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 0.8 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 11.1 | GO:0046323 | glucose import(GO:0046323) |
| 0.1 | 2.7 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.1 | 4.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 4.8 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 1.8 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 2.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.4 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.1 | 12.6 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.1 | 6.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 5.3 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.1 | 5.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.7 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.4 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 8.3 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.1 | 2.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 15.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.3 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 3.5 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 5.9 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 27.7 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.1 | 2.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 2.5 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 10.4 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 5.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 2.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 2.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.9 | GO:1904469 | regulation of tumor necrosis factor secretion(GO:1904467) positive regulation of tumor necrosis factor secretion(GO:1904469) tumor necrosis factor secretion(GO:1990774) |
| 0.1 | 5.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 1.2 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 1.2 | GO:0007100 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.1 | 1.3 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.1 | 0.6 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 1.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 1.6 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 4.0 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.1 | 0.6 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.1 | 0.8 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.5 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.9 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 1.3 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 1.9 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 1.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 2.6 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 3.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 2.4 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.2 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.7 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 1.2 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 | 0.2 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.4 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.0 | 0.4 | GO:0009584 | detection of visible light(GO:0009584) |
| 0.0 | 0.5 | GO:0016266 | O-glycan processing(GO:0016266) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.1 | 56.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 9.1 | 36.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 5.3 | 31.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 5.2 | 83.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 4.7 | 71.1 | GO:0097433 | dense body(GO:0097433) |
| 4.7 | 37.3 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 3.7 | 7.5 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 3.4 | 84.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 3.3 | 52.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 3.1 | 18.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 3.0 | 15.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 2.1 | 14.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 2.0 | 8.2 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 1.9 | 13.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 1.8 | 9.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 1.8 | 26.7 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 1.8 | 8.9 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 1.7 | 13.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.7 | 5.0 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 1.6 | 14.7 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.4 | 4.2 | GO:0098536 | deuterosome(GO:0098536) |
| 1.4 | 72.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 1.4 | 313.1 | GO:0030426 | growth cone(GO:0030426) |
| 1.3 | 20.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 1.2 | 3.7 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 1.1 | 7.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.1 | 8.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.1 | 7.7 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 1.1 | 14.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.1 | 5.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.1 | 51.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 1.0 | 4.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 1.0 | 3.0 | GO:0070701 | mucus layer(GO:0070701) |
| 1.0 | 6.8 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 1.0 | 67.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.9 | 13.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.9 | 8.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.8 | 6.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.7 | 13.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.7 | 2.8 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.7 | 5.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.7 | 6.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.6 | 12.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.6 | 10.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.6 | 18.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.6 | 41.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.6 | 76.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.6 | 3.0 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.6 | 3.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.6 | 14.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.6 | 11.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.5 | 8.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.5 | 14.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.5 | 6.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.5 | 14.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.5 | 5.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.5 | 16.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.4 | 5.8 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.4 | 26.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.4 | 4.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 1.2 | GO:0016939 | plus-end kinesin complex(GO:0005873) kinesin II complex(GO:0016939) |
| 0.4 | 9.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 3.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.4 | 20.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.4 | 55.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.3 | 6.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 10.7 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.3 | 21.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.3 | 2.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 5.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 4.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 2.7 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 18.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 2.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 4.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 4.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 8.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 3.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 21.6 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.2 | 25.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 1.2 | GO:0097504 | SMN complex(GO:0032797) Gemini of coiled bodies(GO:0097504) |
| 0.2 | 2.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 7.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 19.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.2 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 81.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.2 | 1.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 2.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 5.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.2 | 3.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 8.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 11.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 7.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 17.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 3.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 4.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 1.7 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 6.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 4.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 6.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 11.5 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 3.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.6 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 15.3 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.1 | 10.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 4.2 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 2.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 10.0 | GO:0098794 | postsynapse(GO:0098794) |
| 0.1 | 9.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 160.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 2.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 5.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 3.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 28.9 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 4.0 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.1 | 5.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 5.1 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.1 | 4.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 2.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 3.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 2.2 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 3.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 33.2 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 3.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 61.4 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.0 | 79.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 5.5 | 65.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 5.0 | 15.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 4.9 | 29.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 4.9 | 14.7 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 4.9 | 24.3 | GO:0052847 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 4.7 | 61.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 4.6 | 13.8 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 4.5 | 18.0 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 4.4 | 88.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 4.2 | 62.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 3.9 | 19.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 3.8 | 49.8 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 3.7 | 14.9 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 3.7 | 29.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 3.5 | 24.6 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 3.4 | 17.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 3.2 | 38.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) TPR domain binding(GO:0030911) |
| 3.1 | 9.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 3.1 | 9.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 3.1 | 107.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 3.0 | 14.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 2.9 | 14.6 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 2.9 | 11.6 | GO:0097001 | ceramide binding(GO:0097001) |
| 2.9 | 8.6 | GO:0008431 | vitamin E binding(GO:0008431) |
| 2.8 | 8.3 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) sodium:dicarboxylate symporter activity(GO:0017153) |
| 2.8 | 11.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 2.8 | 16.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 2.6 | 23.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 2.5 | 10.0 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 2.5 | 7.5 | GO:0042806 | amidase activity(GO:0004040) fucose binding(GO:0042806) |
| 2.5 | 32.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 2.4 | 31.7 | GO:0008199 | ferric iron binding(GO:0008199) |
| 2.4 | 14.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 2.4 | 7.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 2.3 | 18.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 2.2 | 15.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 2.0 | 8.2 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 2.0 | 7.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 2.0 | 58.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.8 | 10.8 | GO:0008443 | 6-phosphofructo-2-kinase activity(GO:0003873) phosphofructokinase activity(GO:0008443) |
| 1.8 | 26.9 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 1.7 | 6.9 | GO:0035473 | lipase binding(GO:0035473) |
| 1.7 | 5.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 1.7 | 48.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 1.6 | 14.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.6 | 7.8 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 1.5 | 4.6 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 1.5 | 6.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 1.5 | 44.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 1.5 | 7.6 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 1.5 | 7.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 1.4 | 4.3 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 1.4 | 6.9 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 1.4 | 23.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 1.3 | 25.4 | GO:0031005 | filamin binding(GO:0031005) |
| 1.3 | 10.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.3 | 12.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.2 | 3.7 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 1.2 | 5.0 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 1.2 | 15.7 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.2 | 9.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.2 | 6.0 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 1.2 | 5.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.2 | 7.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 1.1 | 3.4 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 1.1 | 6.8 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 1.1 | 7.7 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 1.1 | 6.6 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 1.1 | 8.5 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 1.1 | 14.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.1 | 3.2 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 1.1 | 7.4 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 1.1 | 5.3 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 1.0 | 5.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 1.0 | 3.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.0 | 9.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 1.0 | 4.0 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 1.0 | 8.9 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 1.0 | 7.9 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 1.0 | 2.9 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 1.0 | 5.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.9 | 17.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.9 | 9.1 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.9 | 9.8 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.9 | 3.5 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.9 | 5.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.9 | 6.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.9 | 6.9 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.9 | 17.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.9 | 10.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.8 | 8.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.8 | 5.6 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.8 | 4.0 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.8 | 30.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.8 | 4.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.8 | 13.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.8 | 4.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.7 | 62.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.7 | 7.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.7 | 5.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.7 | 3.5 | GO:0004920 | type I interferon receptor activity(GO:0004905) interleukin-10 receptor activity(GO:0004920) |
| 0.7 | 55.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.7 | 2.0 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.7 | 3.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.7 | 17.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.7 | 24.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.6 | 8.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.6 | 7.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.6 | 3.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.6 | 10.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.6 | 12.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.6 | 3.0 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.6 | 1.8 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.6 | 7.7 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.6 | 1.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.6 | 7.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.6 | 3.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.6 | 8.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.6 | 5.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.5 | 5.9 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.5 | 13.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.5 | 30.5 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.5 | 3.7 | GO:0005497 | androgen binding(GO:0005497) |
| 0.5 | 14.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.5 | 10.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.5 | 7.8 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.5 | 8.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.5 | 2.6 | GO:0004803 | transposase activity(GO:0004803) |
| 0.5 | 4.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.5 | 1.4 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.5 | 9.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.5 | 4.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.5 | 3.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.5 | 1.8 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.4 | 4.0 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 5.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.4 | 2.6 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.4 | 6.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 1.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.4 | 2.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.4 | 14.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.4 | 1.6 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.4 | 8.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.4 | 4.5 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.4 | 3.7 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.4 | 8.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.4 | 5.9 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.4 | 3.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.4 | 2.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.4 | 8.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.4 | 5.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.4 | 4.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.4 | 3.6 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.4 | 5.3 | GO:0043522 | cAMP response element binding protein binding(GO:0008140) leucine zipper domain binding(GO:0043522) |
| 0.4 | 12.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.4 | 19.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.3 | 9.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.3 | 42.0 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.3 | 5.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 6.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 3.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.3 | 1.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.3 | 1.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 5.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 19.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.3 | 3.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.3 | 9.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 9.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 16.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.2 | 0.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.2 | 7.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 2.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 20.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 15.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 6.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 4.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 5.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 2.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 2.6 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.2 | 11.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 3.0 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 0.8 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 0.7 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 6.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 1.4 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.2 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.2 | 0.5 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.2 | 3.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 9.4 | GO:0005230 | extracellular ligand-gated ion channel activity(GO:0005230) |
| 0.2 | 3.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 8.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 17.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.6 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 13.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 2.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 2.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 3.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 22.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.8 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 35.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 9.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 5.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 3.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 3.2 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 2.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 2.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 28.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 6.9 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 3.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 7.2 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 21.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.6 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.1 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 5.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 4.3 | GO:0043492 | ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.1 | 6.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 0.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.8 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 13.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.6 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 3.7 | GO:0001159 | core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 13.1 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 8.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 2.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 111.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 1.6 | 241.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.8 | 35.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.7 | 19.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.6 | 43.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.6 | 2.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.6 | 25.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.5 | 20.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.5 | 8.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.4 | 16.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.4 | 11.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.4 | 32.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.4 | 23.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.4 | 10.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.4 | 20.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.4 | 16.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.4 | 8.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.4 | 7.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 2.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.4 | 7.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.4 | 1.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.4 | 9.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 9.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 25.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.3 | 14.3 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.3 | 11.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.3 | 10.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 3.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.3 | 7.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 11.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 11.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 7.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.2 | 5.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 6.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 1.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 19.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 10.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 9.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 5.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 5.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 4.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 4.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 5.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 3.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 8.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 6.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 4.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 13.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 84.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 1.6 | 44.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 1.4 | 33.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 1.4 | 16.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.2 | 15.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 1.1 | 16.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 1.1 | 14.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 1.0 | 44.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.9 | 34.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.9 | 20.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.8 | 23.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.7 | 20.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.7 | 48.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.7 | 8.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.7 | 12.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.6 | 20.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.6 | 13.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.6 | 44.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.6 | 80.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.6 | 15.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.6 | 9.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.6 | 4.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.5 | 64.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.5 | 9.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.5 | 18.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 16.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.5 | 5.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.4 | 9.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.4 | 8.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.4 | 12.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.4 | 9.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.4 | 22.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 15.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 2.7 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.3 | 4.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 10.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.3 | 7.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 9.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 7.6 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.3 | 17.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.3 | 4.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.3 | 7.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 3.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 6.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.2 | 9.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 4.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 2.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 5.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 6.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 33.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 7.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 2.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 4.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 3.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 33.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 4.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 6.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 5.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 3.5 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.2 | 3.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.2 | 4.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 2.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 5.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 5.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 49.6 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 4.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 10.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 2.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 30.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 4.0 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.1 | 10.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 4.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 4.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 0.7 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 0.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 2.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 0.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 3.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |