Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for OLIG2_NEUROD1_ATOH1
Z-value: 0.91
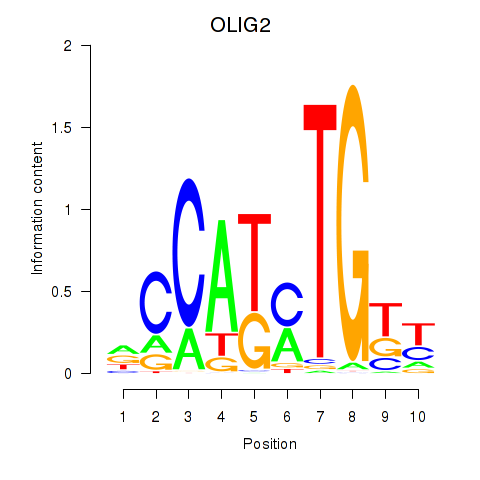
Transcription factors associated with OLIG2_NEUROD1_ATOH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG2
|
ENSG00000205927.5 | OLIG2 |
|
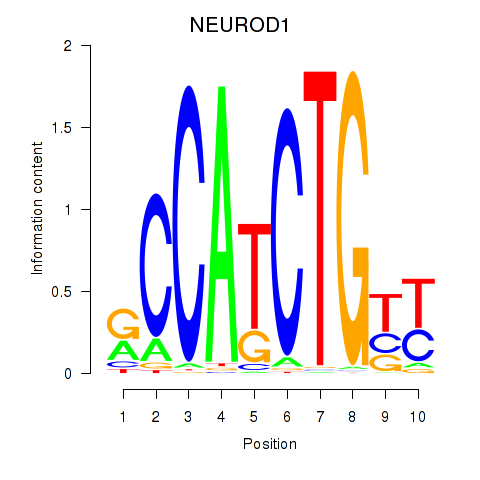
NEUROD1
|
ENSG00000162992.5 | NEUROD1 |
|
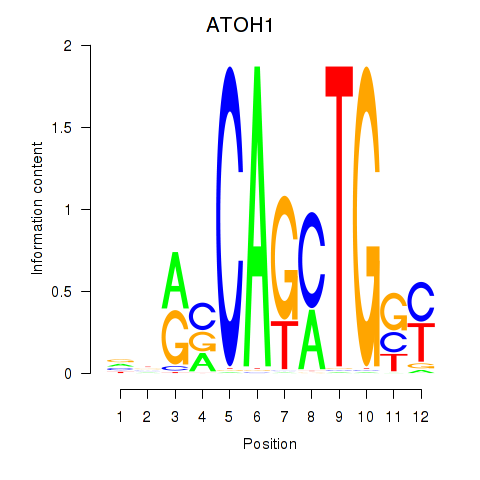
ATOH1
|
ENSG00000172238.6 | ATOH1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATOH1 | hg38_v1_chr4_+_93828746_93828758 | -0.19 | 5.5e-03 | Click! |
| OLIG2 | hg38_v1_chr21_+_33025927_33025942 | 0.10 | 1.3e-01 | Click! |
| NEUROD1 | hg38_v1_chr2_-_181680490_181680525 | 0.06 | 3.8e-01 | Click! |
Activity profile of OLIG2_NEUROD1_ATOH1 motif
Sorted Z-values of OLIG2_NEUROD1_ATOH1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG2_NEUROD1_ATOH1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.9 | 178.8 | GO:0009414 | response to water deprivation(GO:0009414) |
| 7.2 | 28.8 | GO:0001519 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 4.7 | 18.9 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 4.2 | 41.8 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 4.1 | 16.3 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 3.7 | 14.7 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 3.1 | 27.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 2.6 | 10.4 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 2.1 | 14.9 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 2.1 | 14.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.9 | 11.7 | GO:0019255 | UDP-glucuronate biosynthetic process(GO:0006065) glucose 1-phosphate metabolic process(GO:0019255) |
| 1.8 | 5.3 | GO:2000374 | lysosomal membrane organization(GO:0097212) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 1.5 | 18.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 1.5 | 7.6 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 1.3 | 5.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 1.3 | 7.9 | GO:0015853 | adenine transport(GO:0015853) |
| 1.3 | 19.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 1.3 | 15.0 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.2 | 3.5 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 1.0 | 3.9 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 1.0 | 3.8 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.9 | 6.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.8 | 7.6 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.8 | 2.5 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.7 | 0.7 | GO:1904908 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.7 | 6.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.7 | 6.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.7 | 2.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.7 | 5.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.7 | 4.2 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.7 | 2.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.6 | 4.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.6 | 1.9 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.6 | 3.1 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.6 | 2.8 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.5 | 6.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.5 | 6.6 | GO:0070327 | lysine catabolic process(GO:0006554) thyroid hormone transport(GO:0070327) |
| 0.5 | 3.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.5 | 2.5 | GO:0052572 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.4 | 1.8 | GO:1903891 | regulation of ATF6-mediated unfolded protein response(GO:1903891) |
| 0.4 | 3.5 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.4 | 2.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.4 | 1.7 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.4 | 80.0 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.4 | 9.9 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.4 | 9.6 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.4 | 3.9 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.4 | 4.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.4 | 4.3 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.4 | 5.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.4 | 0.7 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.3 | 3.4 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 2.7 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.3 | 4.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.3 | 3.9 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.3 | 8.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.3 | 6.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.3 | 3.7 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.3 | 4.0 | GO:0090179 | establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 2.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.3 | 7.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.3 | 4.2 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.3 | 2.2 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 5.5 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.3 | 3.8 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.3 | 1.6 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.3 | 1.8 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.3 | 1.0 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.3 | 1.0 | GO:1903786 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.2 | 0.7 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.2 | 1.5 | GO:1901977 | negative regulation of cell cycle checkpoint(GO:1901977) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.2 | 0.7 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 4.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 3.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 23.1 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.2 | 3.0 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 14.0 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.2 | 3.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.2 | 2.8 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 13.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 2.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 5.0 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.2 | 3.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 0.6 | GO:0001794 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 0.2 | 0.8 | GO:2000769 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.2 | 1.0 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.2 | 0.6 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 | 5.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 1.4 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.1 | 3.9 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 2.5 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.7 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 2.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 1.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 5.2 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 3.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 1.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 4.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 6.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 3.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.5 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 2.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.8 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.6 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.1 | 1.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 1.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 1.7 | GO:0021756 | striatum development(GO:0021756) |
| 0.1 | 0.5 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.1 | 2.2 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 2.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.5 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 1.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 1.9 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.1 | 5.4 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 1.0 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 1.8 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 1.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.7 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 3.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 7.6 | GO:0001764 | neuron migration(GO:0001764) |
| 0.1 | 1.9 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 1.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.4 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 2.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 5.9 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 0.4 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.9 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 14.5 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.1 | 0.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.1 | GO:1903659 | regulation of interleukin-18 production(GO:0032661) regulation of cytokine activity(GO:0060300) regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 3.4 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) response to interleukin-15(GO:0070672) |
| 0.0 | 3.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 3.9 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 2.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 7.7 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 7.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 3.4 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.8 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 3.0 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 1.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 3.2 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.7 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 1.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.9 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.7 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.0 | 0.8 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 2.0 | GO:0071806 | protein transmembrane transport(GO:0071806) |
| 0.0 | 0.4 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.4 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.8 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 2.4 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 1.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.8 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.5 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.0 | GO:0032364 | oxygen homeostasis(GO:0032364) gas homeostasis(GO:0033483) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 178.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 2.0 | 7.9 | GO:0071817 | MMXD complex(GO:0071817) |
| 1.3 | 6.5 | GO:0031673 | H zone(GO:0031673) |
| 1.3 | 10.3 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 1.0 | 62.3 | GO:0043034 | costamere(GO:0043034) |
| 0.8 | 2.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.6 | 2.4 | GO:0071942 | XPC complex(GO:0071942) |
| 0.6 | 12.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.5 | 2.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.5 | 2.9 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.5 | 1.9 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.5 | 6.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 21.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.4 | 7.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.4 | 4.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 18.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.3 | 3.8 | GO:0072357 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.3 | 1.7 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.3 | 1.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.3 | 10.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.3 | 6.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 2.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 13.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 3.9 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 1.4 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.2 | 3.2 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 10.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 4.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.2 | 0.8 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 3.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 2.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 3.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 43.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 15.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 9.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 1.9 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 3.0 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.8 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 1.0 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 48.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 6.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.5 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 96.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 6.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 2.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 7.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 4.8 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 1.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 14.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 1.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 3.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 11.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 5.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 2.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 4.8 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 3.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 2.6 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 3.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 6.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 23.6 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 2.6 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.0 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.1 | GO:0035102 | PRC1 complex(GO:0035102) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 28.8 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 6.3 | 18.9 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 4.1 | 24.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 2.9 | 11.7 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 2.5 | 54.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 2.1 | 10.4 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 2.0 | 7.9 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 2.0 | 78.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 1.6 | 6.6 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 1.5 | 7.6 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 1.4 | 4.2 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 1.3 | 3.9 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 1.2 | 6.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.2 | 3.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.1 | 12.5 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 1.1 | 19.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 1.0 | 184.0 | GO:0005178 | integrin binding(GO:0005178) |
| 1.0 | 7.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.9 | 52.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.9 | 20.6 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.8 | 8.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.7 | 4.5 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.6 | 3.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.5 | 2.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.5 | 5.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.5 | 2.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.5 | 7.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.4 | 1.7 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.4 | 10.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.4 | 14.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.4 | 4.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 7.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 4.5 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.3 | 8.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 7.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 5.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 12.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 6.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 2.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 1.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 0.8 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.3 | 4.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 4.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 3.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 4.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.8 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 1.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 4.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 1.5 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 2.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 2.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 0.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 0.7 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.2 | 5.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 2.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 13.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.2 | 1.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 3.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 6.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.8 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 3.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 3.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 2.0 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 1.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 1.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 29.6 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 5.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 3.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 2.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 3.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 3.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.4 | GO:0033743 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 1.0 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 1.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 1.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.3 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.1 | 3.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 11.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 3.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.8 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 4.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 3.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 25.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 2.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 2.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 4.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 3.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 4.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 3.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 3.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.8 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.8 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.4 | GO:0016741 | transferase activity, transferring one-carbon groups(GO:0016741) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 202.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.8 | 90.9 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.6 | 11.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 11.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 20.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 2.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 9.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 4.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 3.9 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 4.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 1.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 10.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 4.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 3.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 11.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 5.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 2.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 3.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 169.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 1.1 | 14.9 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.5 | 3.5 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.5 | 11.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.4 | 14.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.4 | 14.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 9.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.3 | 9.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 6.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 3.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 7.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 10.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 4.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 18.9 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.2 | 2.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 6.9 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 4.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 5.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 5.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 2.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 9.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 4.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 3.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 3.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 2.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 5.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 2.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 4.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 5.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 2.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.8 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.5 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.4 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |