|
chr17_-_55511434
Show fit
|
9.81 |
ENST00000636752.1
|
SMIM36
|
small integral membrane protein 36
|
|
chr19_-_55354703
Show fit
|
9.29 |
ENST00000593184.5
ENST00000326529.9
ENST00000589467.1
|
COX6B2
|
cytochrome c oxidase subunit 6B2
|
|
chr7_+_74453790
Show fit
|
6.58 |
ENST00000265755.7
ENST00000424337.7
ENST00000455841.6
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr2_-_61537740
Show fit
|
5.54 |
ENST00000678081.1
ENST00000676889.1
ENST00000677850.1
ENST00000676789.1
|
XPO1
|
exportin 1
|
|
chr8_-_103501890
Show fit
|
5.42 |
ENST00000649416.1
|
ENSG00000285982.1
|
novel protein
|
|
chr3_-_149377637
Show fit
|
4.81 |
ENST00000305366.8
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr2_-_61538180
Show fit
|
4.53 |
ENST00000677150.1
ENST00000678182.1
ENST00000677928.1
ENST00000406957.5
|
XPO1
|
exportin 1
|
|
chr2_-_61538290
Show fit
|
4.43 |
ENST00000678790.1
|
XPO1
|
exportin 1
|
|
chr19_-_55354693
Show fit
|
4.34 |
ENST00000588572.6
|
COX6B2
|
cytochrome c oxidase subunit 6B2
|
|
chr15_+_43800586
Show fit
|
4.19 |
ENST00000442995.4
ENST00000458412.2
|
HYPK
|
huntingtin interacting protein K
|
|
chr8_+_53851786
Show fit
|
4.02 |
ENST00000297313.8
ENST00000344277.10
|
RGS20
|
regulator of G protein signaling 20
|
|
chr13_-_94479671
Show fit
|
3.93 |
ENST00000377028.10
ENST00000446125.1
|
DCT
|
dopachrome tautomerase
|
|
chrX_+_70290077
Show fit
|
3.79 |
ENST00000374403.4
|
KIF4A
|
kinesin family member 4A
|
|
chr11_+_67455352
Show fit
|
3.68 |
ENST00000325656.7
|
CABP4
|
calcium binding protein 4
|
|
chr2_-_224497816
Show fit
|
3.33 |
ENST00000451538.1
|
CUL3
|
cullin 3
|
|
chr8_-_80080816
Show fit
|
3.27 |
ENST00000520527.5
ENST00000517427.5
ENST00000379097.7
ENST00000448733.3
|
TPD52
|
tumor protein D52
|
|
chr12_+_53300027
Show fit
|
3.07 |
ENST00000549488.5
|
MYG1
|
MYG1 exonuclease
|
|
chr10_-_125028061
Show fit
|
3.06 |
ENST00000309035.11
|
CTBP2
|
C-terminal binding protein 2
|
|
chr1_-_165445088
Show fit
|
3.01 |
ENST00000359842.10
|
RXRG
|
retinoid X receptor gamma
|
|
chr2_+_191276885
Show fit
|
2.95 |
ENST00000392316.5
|
MYO1B
|
myosin IB
|
|
chr18_-_66604076
Show fit
|
2.83 |
ENST00000540086.5
ENST00000580157.2
ENST00000262150.7
|
CDH19
|
cadherin 19
|
|
chr1_+_180928133
Show fit
|
2.77 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614
|
|
chr12_+_53299682
Show fit
|
2.46 |
ENST00000267103.10
ENST00000548632.5
|
MYG1
|
MYG1 exonuclease
|
|
chr11_+_102112445
Show fit
|
2.44 |
ENST00000524575.5
|
YAP1
|
Yes1 associated transcriptional regulator
|
|
chr1_+_200894892
Show fit
|
2.41 |
ENST00000413687.3
|
INAVA
|
innate immunity activator
|
|
chrX_+_41447322
Show fit
|
2.32 |
ENST00000378220.2
ENST00000342595.2
|
NYX
|
nyctalopin
|
|
chr15_+_86079863
Show fit
|
2.14 |
ENST00000614907.3
ENST00000441037.7
|
AGBL1
|
ATP/GTP binding protein like 1
|
|
chr1_-_150235995
Show fit
|
2.11 |
ENST00000436748.6
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E
|
|
chr11_-_11353241
Show fit
|
2.09 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3
|
|
chr1_-_150236064
Show fit
|
2.03 |
ENST00000532744.2
ENST00000369114.9
ENST00000369115.3
ENST00000583931.6
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E
|
|
chr1_-_150236150
Show fit
|
1.95 |
ENST00000629042.2
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E
|
|
chr8_+_60678705
Show fit
|
1.91 |
ENST00000423902.7
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr4_-_101347327
Show fit
|
1.90 |
ENST00000394853.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha
|
|
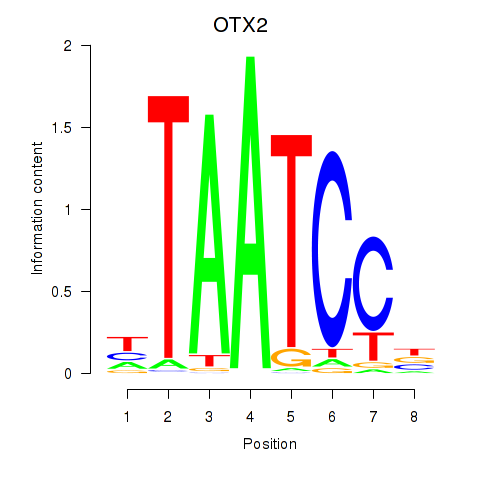
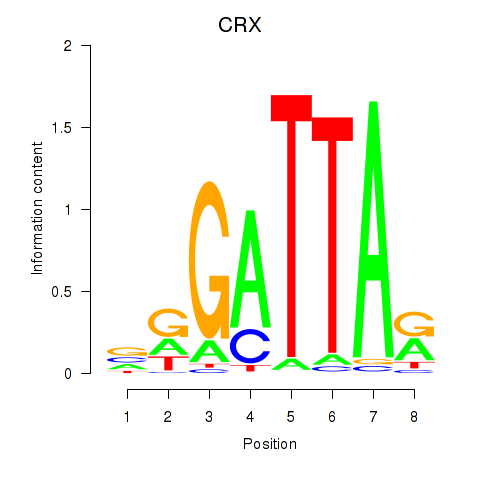
chr19_+_47821907
Show fit
|
1.88 |
ENST00000539067.5
ENST00000221996.12
ENST00000613299.1
|
CRX
|
cone-rod homeobox
|
|
chr4_-_101347492
Show fit
|
1.81 |
ENST00000394854.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha
|
|
chr1_-_165445220
Show fit
|
1.79 |
ENST00000619224.1
|
RXRG
|
retinoid X receptor gamma
|
|
chrX_-_10620419
Show fit
|
1.76 |
ENST00000380782.6
|
MID1
|
midline 1
|
|
chr13_+_73058993
Show fit
|
1.75 |
ENST00000377687.6
|
KLF5
|
Kruppel like factor 5
|
|
chrX_-_54357993
Show fit
|
1.73 |
ENST00000375169.7
ENST00000354646.6
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chr4_-_101347471
Show fit
|
1.73 |
ENST00000323055.10
ENST00000512215.5
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha
|
|
chr2_-_174846405
Show fit
|
1.68 |
ENST00000409597.5
ENST00000413882.6
|
CHN1
|
chimerin 1
|
|
chr3_-_49021045
Show fit
|
1.68 |
ENST00000440857.5
|
DALRD3
|
DALR anticodon binding domain containing 3
|
|
chr11_-_67523396
Show fit
|
1.63 |
ENST00000353903.9
ENST00000294288.5
|
CABP2
|
calcium binding protein 2
|
|
chr4_-_73223082
Show fit
|
1.62 |
ENST00000509867.6
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr20_-_50115935
Show fit
|
1.61 |
ENST00000340309.7
ENST00000415862.6
ENST00000371677.7
|
UBE2V1
|
ubiquitin conjugating enzyme E2 V1
|
|
chr1_-_150235943
Show fit
|
1.61 |
ENST00000533654.5
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E
|
|
chr9_-_128204218
Show fit
|
1.56 |
ENST00000634901.1
ENST00000372948.7
|
CIZ1
|
CDKN1A interacting zinc finger protein 1
|
|
chr12_-_52777343
Show fit
|
1.55 |
ENST00000332411.2
|
KRT76
|
keratin 76
|
|
chr8_-_140718661
Show fit
|
1.49 |
ENST00000430260.6
|
PTK2
|
protein tyrosine kinase 2
|
|
chr15_+_43593054
Show fit
|
1.46 |
ENST00000453782.5
ENST00000300283.10
ENST00000437924.5
|
CKMT1B
|
creatine kinase, mitochondrial 1B
|
|
chr5_-_88883701
Show fit
|
1.44 |
ENST00000636998.1
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr3_-_146528750
Show fit
|
1.43 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1
|
|
chr19_-_51034892
Show fit
|
1.41 |
ENST00000319590.8
ENST00000250351.4
|
KLK12
|
kallikrein related peptidase 12
|
|
chrX_+_46912276
Show fit
|
1.40 |
ENST00000424392.5
ENST00000611250.4
|
JADE3
|
jade family PHD finger 3
|
|
chr4_-_152679984
Show fit
|
1.38 |
ENST00000304385.8
ENST00000504064.1
|
TMEM154
|
transmembrane protein 154
|
|
chr22_-_30471986
Show fit
|
1.37 |
ENST00000401751.5
ENST00000402286.5
ENST00000403066.5
ENST00000215812.9
|
SEC14L3
|
SEC14 like lipid binding 3
|
|
chr4_-_101346842
Show fit
|
1.36 |
ENST00000507176.5
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha
|
|
chr18_+_34710307
Show fit
|
1.35 |
ENST00000679796.1
|
DTNA
|
dystrobrevin alpha
|
|
chr2_-_65432591
Show fit
|
1.35 |
ENST00000356388.9
|
SPRED2
|
sprouty related EVH1 domain containing 2
|
|
chr18_-_24311495
Show fit
|
1.33 |
ENST00000357041.8
|
OSBPL1A
|
oxysterol binding protein like 1A
|
|
chr13_-_30617500
Show fit
|
1.32 |
ENST00000405805.5
|
HMGB1
|
high mobility group box 1
|
|
chrX_+_46912412
Show fit
|
1.30 |
ENST00000614628.5
|
JADE3
|
jade family PHD finger 3
|
|
chr5_-_88883147
Show fit
|
1.26 |
ENST00000513252.5
ENST00000506554.5
ENST00000508569.5
ENST00000637732.1
ENST00000504921.7
ENST00000637481.1
ENST00000510942.5
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr9_-_14300231
Show fit
|
1.22 |
ENST00000636735.1
|
NFIB
|
nuclear factor I B
|
|
chr5_-_150758497
Show fit
|
1.22 |
ENST00000521533.1
ENST00000424236.5
|
DCTN4
|
dynactin subunit 4
|
|
chr10_+_116590956
Show fit
|
1.19 |
ENST00000358834.9
ENST00000528052.5
|
PNLIPRP1
|
pancreatic lipase related protein 1
|
|
chrX_+_101391202
Show fit
|
1.19 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a
|
|
chrX_-_10620534
Show fit
|
1.14 |
ENST00000317552.9
|
MID1
|
midline 1
|
|
chr12_-_52796110
Show fit
|
1.13 |
ENST00000417996.2
|
KRT3
|
keratin 3
|
|
chrX_+_101390976
Show fit
|
1.11 |
ENST00000392994.7
|
RPL36A
|
ribosomal protein L36a
|
|
chr2_-_174847015
Show fit
|
1.10 |
ENST00000650938.1
|
CHN1
|
chimerin 1
|
|
chr12_+_14973020
Show fit
|
1.07 |
ENST00000266395.3
|
PDE6H
|
phosphodiesterase 6H
|
|
chr1_-_150235972
Show fit
|
1.04 |
ENST00000534220.1
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E
|
|
chrX_+_101391000
Show fit
|
1.03 |
ENST00000553110.8
ENST00000409338.5
ENST00000409170.3
|
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a
RPL36A-HNRNPH2 readthrough
|
|
chr5_-_88883199
Show fit
|
1.01 |
ENST00000514015.5
ENST00000503075.1
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr13_+_35476740
Show fit
|
1.00 |
ENST00000537702.5
|
NBEA
|
neurobeachin
|
|
chrX_+_101390824
Show fit
|
1.00 |
ENST00000427805.6
ENST00000614077.4
|
RPL36A
|
ribosomal protein L36a
|
|
chr20_-_35742207
Show fit
|
0.96 |
ENST00000397370.3
ENST00000528062.7
ENST00000374038.7
ENST00000253363.11
ENST00000361162.10
|
RBM39
|
RNA binding motif protein 39
|
|
chr1_-_151992571
Show fit
|
0.96 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10
|
|
chr4_-_110636963
Show fit
|
0.94 |
ENST00000394595.8
|
PITX2
|
paired like homeodomain 2
|
|
chr1_-_173205543
Show fit
|
0.94 |
ENST00000367718.5
|
TNFSF4
|
TNF superfamily member 4
|
|
chr1_-_148152272
Show fit
|
0.92 |
ENST00000682118.1
ENST00000615281.4
|
NBPF11
|
NBPF member 11
|
|
chr10_+_116591010
Show fit
|
0.91 |
ENST00000530319.5
ENST00000527980.5
ENST00000471549.5
ENST00000534537.5
|
PNLIPRP1
|
pancreatic lipase related protein 1
|
|
chrX_-_6228835
Show fit
|
0.88 |
ENST00000381095.8
|
NLGN4X
|
neuroligin 4 X-linked
|
|
chr22_+_25219633
Show fit
|
0.88 |
ENST00000398215.3
|
CRYBB2
|
crystallin beta B2
|
|
chr15_-_26939518
Show fit
|
0.87 |
ENST00000541819.6
|
GABRB3
|
gamma-aminobutyric acid type A receptor subunit beta3
|
|
chr1_+_86914616
Show fit
|
0.86 |
ENST00000370550.10
ENST00000370551.8
|
HS2ST1
|
heparan sulfate 2-O-sulfotransferase 1
|
|
chrX_+_28587411
Show fit
|
0.86 |
ENST00000378993.6
|
IL1RAPL1
|
interleukin 1 receptor accessory protein like 1
|
|
chr10_+_102132994
Show fit
|
0.85 |
ENST00000413464.6
ENST00000278070.7
|
PPRC1
|
PPARG related coactivator 1
|
|
chr12_-_89656093
Show fit
|
0.85 |
ENST00000359142.7
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1
|
|
chr4_+_128809684
Show fit
|
0.85 |
ENST00000226319.11
ENST00000511647.5
|
JADE1
|
jade family PHD finger 1
|
|
chr10_-_63269057
Show fit
|
0.82 |
ENST00000542921.5
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr12_-_89656051
Show fit
|
0.82 |
ENST00000261173.6
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1
|
|
chr6_-_106975616
Show fit
|
0.81 |
ENST00000610952.1
|
CD24
|
CD24 molecule
|
|
chr4_+_128809791
Show fit
|
0.80 |
ENST00000452328.6
ENST00000504089.5
|
JADE1
|
jade family PHD finger 1
|
|
chr14_-_34630109
Show fit
|
0.79 |
ENST00000396526.7
|
SNX6
|
sorting nexin 6
|
|
chr10_+_94089034
Show fit
|
0.79 |
ENST00000676102.1
ENST00000371385.8
|
PLCE1
|
phospholipase C epsilon 1
|
|
chr9_+_12693327
Show fit
|
0.79 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1
|
|
chr22_-_37984534
Show fit
|
0.78 |
ENST00000396884.8
|
SOX10
|
SRY-box transcription factor 10
|
|
chr10_+_94089067
Show fit
|
0.78 |
ENST00000371375.1
ENST00000675218.1
|
PLCE1
|
phospholipase C epsilon 1
|
|
chr15_+_33310993
Show fit
|
0.76 |
ENST00000634418.1
ENST00000634750.1
|
RYR3
|
ryanodine receptor 3
|
|
chr17_-_40703744
Show fit
|
0.76 |
ENST00000264651.3
|
KRT24
|
keratin 24
|
|
chr4_+_125314918
Show fit
|
0.76 |
ENST00000674496.2
ENST00000394329.9
|
FAT4
|
FAT atypical cadherin 4
|
|
chr1_+_11691688
Show fit
|
0.76 |
ENST00000294485.6
|
DRAXIN
|
dorsal inhibitory axon guidance protein
|
|
chr3_+_173584433
Show fit
|
0.74 |
ENST00000361589.8
|
NLGN1
|
neuroligin 1
|
|
chr12_-_13095664
Show fit
|
0.74 |
ENST00000337630.10
ENST00000545699.1
|
GSG1
|
germ cell associated 1
|
|
chr1_+_81306096
Show fit
|
0.71 |
ENST00000370721.5
ENST00000370727.5
ENST00000370725.5
ENST00000370723.5
ENST00000370728.5
ENST00000370730.5
|
ADGRL2
|
adhesion G protein-coupled receptor L2
|
|
chr7_+_136869077
Show fit
|
0.70 |
ENST00000320658.9
ENST00000453373.5
ENST00000401861.1
|
CHRM2
|
cholinergic receptor muscarinic 2
|
|
chrX_-_15600953
Show fit
|
0.70 |
ENST00000679212.1
ENST00000679278.1
ENST00000678046.1
ENST00000252519.8
|
ACE2
|
angiotensin I converting enzyme 2
|
|
chr10_-_125161019
Show fit
|
0.70 |
ENST00000411419.6
|
CTBP2
|
C-terminal binding protein 2
|
|
chr19_-_3772211
Show fit
|
0.69 |
ENST00000555978.5
ENST00000555633.3
|
RAX2
|
retina and anterior neural fold homeobox 2
|
|
chr1_+_167329044
Show fit
|
0.69 |
ENST00000367862.9
|
POU2F1
|
POU class 2 homeobox 1
|
|
chrX_-_102516714
Show fit
|
0.69 |
ENST00000289373.5
|
TMSB15A
|
thymosin beta 15A
|
|
chr19_+_39498938
Show fit
|
0.68 |
ENST00000356433.10
ENST00000596614.5
ENST00000205143.4
|
DLL3
|
delta like canonical Notch ligand 3
|
|
chr2_-_70553440
Show fit
|
0.67 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha
|
|
chr10_+_68901258
Show fit
|
0.67 |
ENST00000373585.8
|
DDX50
|
DExD-box helicase 50
|
|
chr16_-_31064952
Show fit
|
0.67 |
ENST00000426488.6
|
ZNF668
|
zinc finger protein 668
|
|
chr9_-_115118198
Show fit
|
0.66 |
ENST00000534839.1
ENST00000535648.5
|
TNC
|
tenascin C
|
|
chr3_-_65597886
Show fit
|
0.65 |
ENST00000460329.6
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr12_-_14696571
Show fit
|
0.65 |
ENST00000261170.5
|
GUCY2C
|
guanylate cyclase 2C
|
|
chr10_-_110304894
Show fit
|
0.65 |
ENST00000369603.10
|
SMNDC1
|
survival motor neuron domain containing 1
|
|
chr5_+_51383394
Show fit
|
0.64 |
ENST00000230658.12
|
ISL1
|
ISL LIM homeobox 1
|
|
chr1_-_223845894
Show fit
|
0.63 |
ENST00000391878.6
ENST00000343537.12
|
TP53BP2
|
tumor protein p53 binding protein 2
|
|
chr2_-_47906437
Show fit
|
0.62 |
ENST00000403359.8
|
FBXO11
|
F-box protein 11
|
|
chr3_+_130931893
Show fit
|
0.60 |
ENST00000504612.5
|
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1
|
|
chrX_-_15601077
Show fit
|
0.58 |
ENST00000680121.1
|
ACE2
|
angiotensin I converting enzyme 2
|
|
chr12_-_13095798
Show fit
|
0.58 |
ENST00000396302.7
|
GSG1
|
germ cell associated 1
|
|
chr22_-_19150292
Show fit
|
0.58 |
ENST00000086933.3
|
GSC2
|
goosecoid homeobox 2
|
|
chr15_+_79311084
Show fit
|
0.57 |
ENST00000299705.10
|
TMED3
|
transmembrane p24 trafficking protein 3
|
|
chr3_-_197226351
Show fit
|
0.56 |
ENST00000656428.1
|
DLG1
|
discs large MAGUK scaffold protein 1
|
|
chr2_+_161624406
Show fit
|
0.56 |
ENST00000446997.6
ENST00000272716.9
|
SLC4A10
|
solute carrier family 4 member 10
|
|
chr21_-_33542841
Show fit
|
0.56 |
ENST00000381831.7
ENST00000381839.7
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase
|
|
chr6_-_111483700
Show fit
|
0.56 |
ENST00000435970.5
ENST00000358835.7
|
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit
|
|
chr11_-_30586866
Show fit
|
0.55 |
ENST00000528686.2
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr5_-_77776261
Show fit
|
0.55 |
ENST00000518338.6
ENST00000520039.1
ENST00000306388.10
ENST00000520361.5
ENST00000651106.1
|
TBCA
|
tubulin folding cofactor A
|
|
chr18_-_31102411
Show fit
|
0.55 |
ENST00000251081.8
ENST00000280904.11
ENST00000682357.1
ENST00000648081.1
|
DSC2
|
desmocollin 2
|
|
chr12_-_13095628
Show fit
|
0.55 |
ENST00000457134.6
ENST00000537302.5
|
GSG1
|
germ cell associated 1
|
|
chr9_+_69145463
Show fit
|
0.53 |
ENST00000636438.1
|
TJP2
|
tight junction protein 2
|
|
chrY_+_14522573
Show fit
|
0.53 |
ENST00000643089.1
ENST00000382872.5
|
NLGN4Y
|
neuroligin 4 Y-linked
|
|
chr15_-_31161157
Show fit
|
0.53 |
ENST00000542188.5
|
TRPM1
|
transient receptor potential cation channel subfamily M member 1
|
|
chr16_-_31065011
Show fit
|
0.52 |
ENST00000539836.3
ENST00000535577.5
ENST00000442862.2
|
ZNF668
|
zinc finger protein 668
|
|
chr11_-_62984957
Show fit
|
0.52 |
ENST00000377871.7
ENST00000360421.9
|
SLC22A6
|
solute carrier family 22 member 6
|
|
chr8_-_90082871
Show fit
|
0.51 |
ENST00000265431.7
|
CALB1
|
calbindin 1
|
|
chr21_-_18403754
Show fit
|
0.51 |
ENST00000284885.8
|
TMPRSS15
|
transmembrane serine protease 15
|
|
chr2_-_55010348
Show fit
|
0.49 |
ENST00000394609.6
|
RTN4
|
reticulon 4
|
|
chr17_+_58148384
Show fit
|
0.48 |
ENST00000268912.6
ENST00000641449.1
|
OR4D1
|
olfactory receptor family 4 subfamily D member 1
|
|
chr17_-_41505597
Show fit
|
0.47 |
ENST00000336861.7
ENST00000246635.8
ENST00000587544.5
ENST00000587435.1
|
KRT13
|
keratin 13
|
|
chr3_+_49021071
Show fit
|
0.46 |
ENST00000395458.6
|
NDUFAF3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3
|
|
chr19_-_51034727
Show fit
|
0.46 |
ENST00000525263.5
|
KLK12
|
kallikrein related peptidase 12
|
|
chr5_-_88883420
Show fit
|
0.45 |
ENST00000437473.6
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_+_153222307
Show fit
|
0.44 |
ENST00000675899.1
ENST00000675611.1
ENST00000674872.1
ENST00000676167.1
|
TRIM2
|
tripartite motif containing 2
|
|
chr2_-_144431001
Show fit
|
0.44 |
ENST00000636413.1
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr3_+_69866217
Show fit
|
0.43 |
ENST00000314589.10
|
MITF
|
melanocyte inducing transcription factor
|
|
chr7_+_136868622
Show fit
|
0.42 |
ENST00000680005.1
ENST00000445907.6
|
CHRM2
|
cholinergic receptor muscarinic 2
|
|
chr9_+_85941121
Show fit
|
0.42 |
ENST00000361671.10
|
NAA35
|
N-alpha-acetyltransferase 35, NatC auxiliary subunit
|
|
chr6_+_12716801
Show fit
|
0.41 |
ENST00000674595.1
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr15_+_96325935
Show fit
|
0.41 |
ENST00000421109.6
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2
|
|
chrX_-_124963768
Show fit
|
0.40 |
ENST00000371130.7
ENST00000422452.2
|
TENM1
|
teneurin transmembrane protein 1
|
|
chr17_+_62458641
Show fit
|
0.40 |
ENST00000582809.5
|
TLK2
|
tousled like kinase 2
|
|
chr4_+_67558719
Show fit
|
0.39 |
ENST00000265404.7
ENST00000396225.1
|
STAP1
|
signal transducing adaptor family member 1
|
|
chr4_+_153204410
Show fit
|
0.39 |
ENST00000675838.1
ENST00000674967.1
ENST00000632856.2
ENST00000441616.6
ENST00000433687.2
ENST00000494872.6
ENST00000460908.2
ENST00000675780.1
ENST00000674976.1
ENST00000338700.10
ENST00000675293.1
ENST00000676172.1
ENST00000675673.1
ENST00000675492.1
ENST00000675425.1
ENST00000675384.1
ENST00000675063.1
ENST00000675340.1
ENST00000675835.1
ENST00000675054.1
ENST00000675710.1
ENST00000502281.3
|
ENSG00000288637.1
TRIM2
|
novel protein
tripartite motif containing 2
|
|
chr11_-_33717409
Show fit
|
0.39 |
ENST00000651485.1
|
CD59
|
CD59 molecule (CD59 blood group)
|
|
chr1_+_35268663
Show fit
|
0.38 |
ENST00000314607.11
|
ZMYM4
|
zinc finger MYM-type containing 4
|
|
chr5_-_142325001
Show fit
|
0.37 |
ENST00000344120.4
ENST00000434127.3
|
SPRY4
|
sprouty RTK signaling antagonist 4
|
|
chrX_+_12137409
Show fit
|
0.36 |
ENST00000672010.1
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr17_+_77281429
Show fit
|
0.36 |
ENST00000591198.5
ENST00000427177.6
|
SEPTIN9
|
septin 9
|
|
chrX_+_70268305
Show fit
|
0.36 |
ENST00000374495.7
|
ARR3
|
arrestin 3
|
|
chr8_-_99893135
Show fit
|
0.36 |
ENST00000524245.5
|
COX6C
|
cytochrome c oxidase subunit 6C
|
|
chr2_+_169509693
Show fit
|
0.36 |
ENST00000284669.2
|
KLHL41
|
kelch like family member 41
|
|
chr3_+_49020443
Show fit
|
0.35 |
ENST00000326912.8
|
NDUFAF3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3
|
|
chr9_+_100442271
Show fit
|
0.35 |
ENST00000502978.1
|
MSANTD3-TMEFF1
|
MSANTD3-TMEFF1 readthrough
|
|
chr10_+_22321056
Show fit
|
0.34 |
ENST00000376663.8
|
BMI1
|
BMI1 proto-oncogene, polycomb ring finger
|
|
chr15_-_52191387
Show fit
|
0.34 |
ENST00000261837.12
|
GNB5
|
G protein subunit beta 5
|
|
chr14_-_36320582
Show fit
|
0.34 |
ENST00000604336.5
ENST00000359527.11
ENST00000621657.4
ENST00000603139.1
ENST00000318473.11
ENST00000416007.9
|
MBIP
|
MAP3K12 binding inhibitory protein 1
|
|
chr1_-_186461089
Show fit
|
0.34 |
ENST00000391997.3
|
PDC
|
phosducin
|
|
chr17_+_8002610
Show fit
|
0.34 |
ENST00000254854.5
|
GUCY2D
|
guanylate cyclase 2D, retinal
|
|
chr7_-_105269007
Show fit
|
0.33 |
ENST00000357311.7
|
SRPK2
|
SRSF protein kinase 2
|
|
chr6_+_42173358
Show fit
|
0.33 |
ENST00000372958.2
|
GUCA1A
|
guanylate cyclase activator 1A
|
|
chr5_+_162067764
Show fit
|
0.32 |
ENST00000639213.2
ENST00000414552.6
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2
|
|
chr7_-_78771108
Show fit
|
0.32 |
ENST00000626691.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr5_-_77776321
Show fit
|
0.32 |
ENST00000380377.9
|
TBCA
|
tubulin folding cofactor A
|
|
chr19_-_51034840
Show fit
|
0.30 |
ENST00000529888.5
|
KLK12
|
kallikrein related peptidase 12
|
|
chr6_+_12716554
Show fit
|
0.29 |
ENST00000676159.1
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr5_-_150289941
Show fit
|
0.29 |
ENST00000682786.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha
|
|
chr15_+_33310946
Show fit
|
0.28 |
ENST00000415757.7
ENST00000634891.2
ENST00000389232.9
ENST00000622037.1
|
RYR3
|
ryanodine receptor 3
|
|
chr12_-_86256299
Show fit
|
0.27 |
ENST00000552808.6
ENST00000547225.5
|
MGAT4C
|
MGAT4 family member C
|
|
chr12_-_66803980
Show fit
|
0.27 |
ENST00000539540.5
ENST00000540433.5
ENST00000541947.1
ENST00000538373.1
|
GRIP1
|
glutamate receptor interacting protein 1
|
|
chrX_+_130339941
Show fit
|
0.26 |
ENST00000218197.9
|
SLC25A14
|
solute carrier family 25 member 14
|
|
chr10_-_61001430
Show fit
|
0.26 |
ENST00000357917.4
|
RHOBTB1
|
Rho related BTB domain containing 1
|
|
chr12_-_91058016
Show fit
|
0.26 |
ENST00000266719.4
|
KERA
|
keratocan
|
|
chr12_-_56454653
Show fit
|
0.26 |
ENST00000652304.1
|
MIP
|
major intrinsic protein of lens fiber
|
|
chr12_+_77830886
Show fit
|
0.24 |
ENST00000397909.7
ENST00000549464.5
|
NAV3
|
neuron navigator 3
|
|
chr5_-_150289625
Show fit
|
0.23 |
ENST00000683332.1
ENST00000398376.8
ENST00000672785.1
ENST00000672396.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha
|
|
chr2_-_212124901
Show fit
|
0.23 |
ENST00000402597.6
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4
|
|
chr15_+_79311137
Show fit
|
0.23 |
ENST00000424155.6
ENST00000536821.5
|
TMED3
|
transmembrane p24 trafficking protein 3
|
|
chr2_+_172928165
Show fit
|
0.23 |
ENST00000535187.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4
|
|
chr5_+_162067458
Show fit
|
0.23 |
ENST00000639975.1
ENST00000639111.2
ENST00000639683.1
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2
|
|
chr5_-_33984681
Show fit
|
0.23 |
ENST00000296589.9
|
SLC45A2
|
solute carrier family 45 member 2
|
|
chr17_+_62479025
Show fit
|
0.22 |
ENST00000326270.13
ENST00000682203.1
ENST00000343388.11
ENST00000683536.1
ENST00000684440.1
ENST00000684772.1
ENST00000682075.1
ENST00000683104.1
ENST00000684012.1
|
TLK2
|
tousled like kinase 2
|
|
chr14_+_58427686
Show fit
|
0.22 |
ENST00000650904.1
ENST00000652326.2
ENST00000554463.5
ENST00000555833.5
|
KIAA0586
|
KIAA0586
|