Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
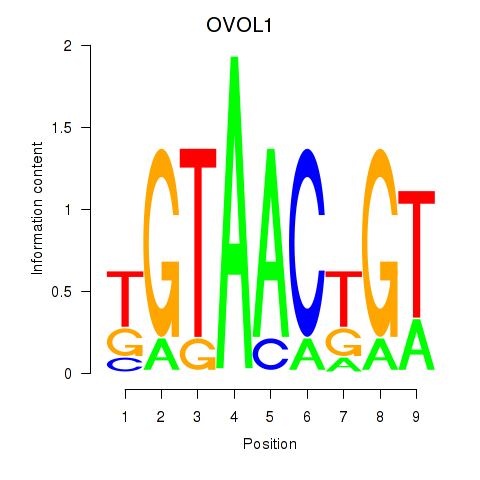
Results for OVOL1
Z-value: 1.51
Transcription factors associated with OVOL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OVOL1
|
ENSG00000172818.10 | OVOL1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OVOL1 | hg38_v1_chr11_+_65787056_65787075 | 0.18 | 8.3e-03 | Click! |
Activity profile of OVOL1 motif
Sorted Z-values of OVOL1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of OVOL1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_83709382 | 10.91 |
ENST00000520302.5
ENST00000520213.5 ENST00000439399.6 |
SNAP91
|
synaptosome associated protein 91 |
| chr11_+_123430259 | 10.44 |
ENST00000533341.3
ENST00000635736.2 |
GRAMD1B
|
GRAM domain containing 1B |
| chr18_-_5543960 | 8.88 |
ENST00000400111.8
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr18_-_5544250 | 8.65 |
ENST00000540638.6
ENST00000342933.7 |
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr18_-_5543988 | 8.62 |
ENST00000341928.7
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr5_+_161850597 | 7.76 |
ENST00000634335.1
ENST00000635880.1 |
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr5_+_141359970 | 7.52 |
ENST00000522605.2
ENST00000622527.1 |
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr5_-_16508788 | 7.19 |
ENST00000682142.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr10_-_72088972 | 7.08 |
ENST00000317376.8
ENST00000412663.5 |
SPOCK2
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 2 |
| chr5_-_16508990 | 6.98 |
ENST00000399793.6
|
RETREG1
|
reticulophagy regulator 1 |
| chr10_-_72088533 | 6.72 |
ENST00000373109.7
|
SPOCK2
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 2 |
| chr19_-_49442360 | 6.16 |
ENST00000600601.5
|
SLC17A7
|
solute carrier family 17 member 7 |
| chr5_-_16508812 | 6.02 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr5_-_16508858 | 5.80 |
ENST00000684456.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr5_-_16508951 | 5.67 |
ENST00000682628.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr5_+_140882116 | 4.56 |
ENST00000289272.3
ENST00000409494.5 ENST00000617769.1 |
PCDHA13
|
protocadherin alpha 13 |
| chr1_+_174877430 | 2.94 |
ENST00000392064.6
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr19_-_45792755 | 1.25 |
ENST00000377735.7
ENST00000270223.7 |
DMWD
|
DM1 locus, WD repeat containing |
| chr2_-_182522556 | 1.23 |
ENST00000435564.5
|
PDE1A
|
phosphodiesterase 1A |
| chr4_+_39406917 | 1.09 |
ENST00000257408.5
|
KLB
|
klotho beta |
| chr2_-_182522703 | 0.96 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A |
| chr2_-_152099155 | 0.89 |
ENST00000637309.1
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr17_-_7242156 | 0.73 |
ENST00000571129.5
ENST00000571253.1 ENST00000573928.1 ENST00000302386.10 |
GABARAP
|
GABA type A receptor-associated protein |
| chr3_-_165837412 | 0.65 |
ENST00000479451.5
ENST00000488954.1 ENST00000264381.8 |
BCHE
|
butyrylcholinesterase |
| chr18_+_54269493 | 0.62 |
ENST00000579534.6
ENST00000406285.7 ENST00000577612.5 ENST00000579434.5 ENST00000583136.5 |
POLI
|
DNA polymerase iota |
| chr2_-_152099023 | 0.42 |
ENST00000201943.10
ENST00000427385.6 ENST00000539935.7 |
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr15_+_75043263 | 0.11 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 31.7 | GO:0061709 | reticulophagy(GO:0061709) |
| 2.9 | 26.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 2.1 | 6.2 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.8 | 13.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 10.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.3 | 7.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 0.6 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 1.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 2.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 12.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 2.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 26.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.8 | 6.2 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.5 | 31.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 7.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 10.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 13.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.9 | 7.8 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.6 | 13.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 10.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.4 | 2.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 1.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 0.6 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.1 | 26.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 12.1 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 6.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 2.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.1 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |